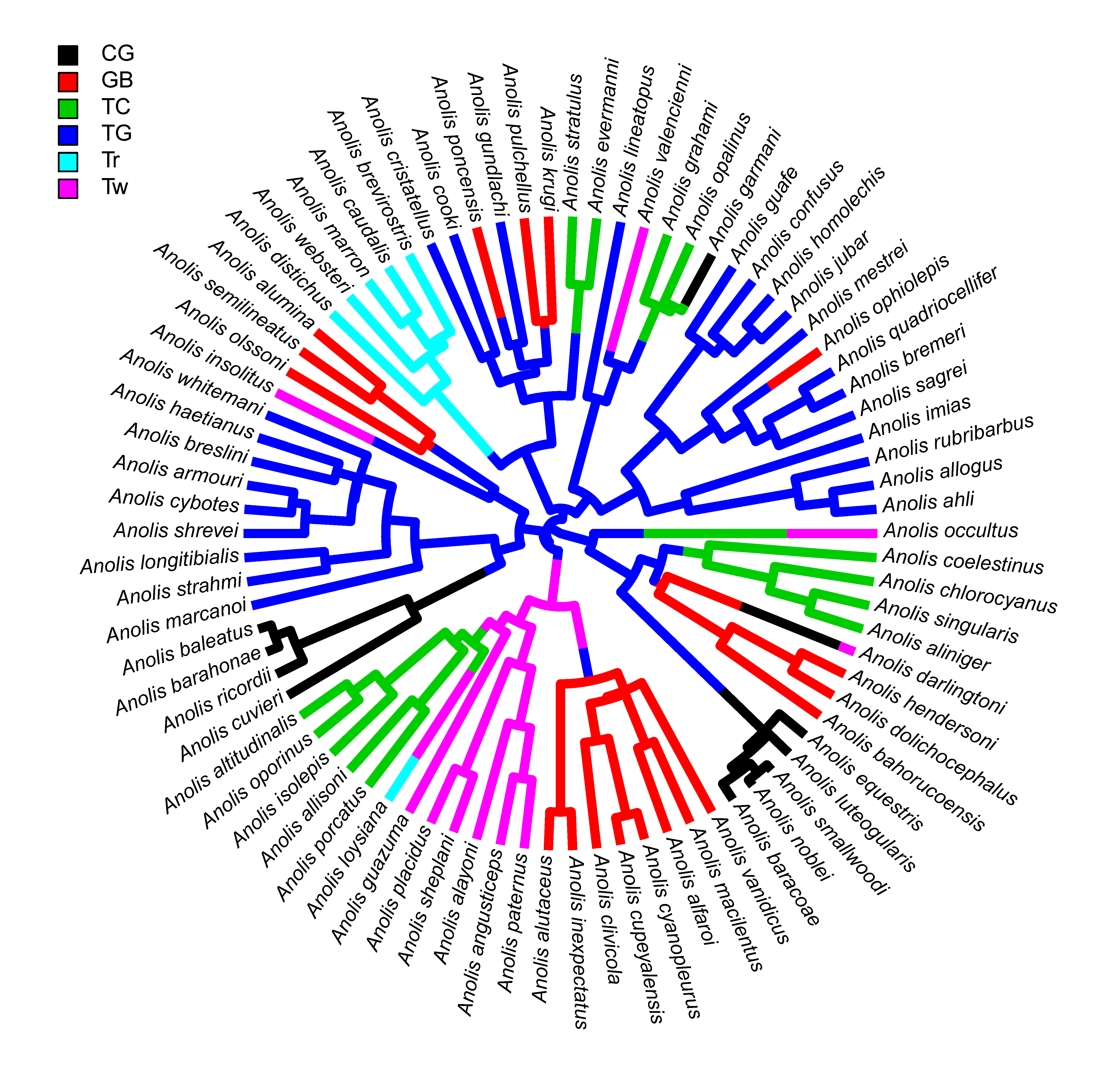
I just pushed a series of updates (1, 2, 3, 4) to fix some issues raised by the authors of ape and phangorn in advance of the submission of new CRAN versions of those two packages.
The first was that I had inadvertently corrupted an S3 method
multi2di of ape when I created the function
multi2di.simmap without exporting a method to the namespace. The
other issues came up in R CMD check and included such things
as failing to import various functions from dependencies, other problems with
the namespace, errors in documenting various variables, the absence of a manual
page for one function (likSurface.rateshift) that is exported
by phytools, and a few problems with the R code of some new functions.
I too hope to update the version of phytools on CRAN as the current version dates to June.
The latest phytools version can be installed from GitHub using devtools as follows:
library(devtools)
install_github("liamrevell/phytools")
Since both phangorn & ape are in the process of being updated, but phytools depends on the latest version of neither, you may need to do:
install_github("liamrevell/phytools",upgrade_dependencies=FALSE)
That's it.