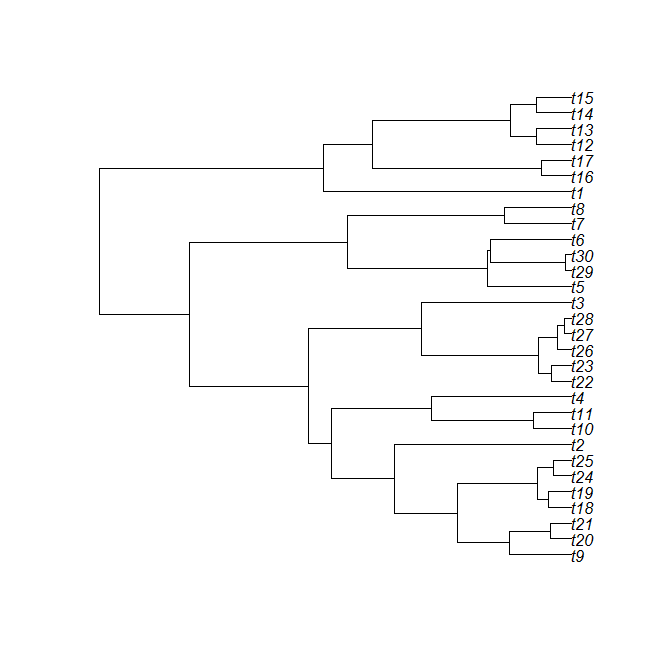
A user asks the following:
I want to plot vertical lines indicating various time points on my tree. . . . Is there an easy way to do this in R. . . ?
The answer turns out to be that yes - it is relatively easy to do this using the base function lines. Let's try:
First, let's use pbtree to simulate a tree and plot.phylo to plot it:
> tree<-pbtree(n=30,scale=100)
> x<-plot(tree)
Ok, now, the dimensions of the plotted area in plot.phylo are 0 to the tree height (plus some extra space for the labels) on the horizontal axis; and 1 through the number of tips (here 30) on the vertical axis. In case we didn't know this already, it is also returned silently by plot.phylo. Here, we have stored this in x:
> x
$type
[1] "phylogram"
...
$x.lim
[1] 0.0000 108.3092
$y.lim
[1] 1 30
...
$Nnode
[1] 29
Now let's add lines at 25, 50, and 75 (time units) above the root:
> # with our plotting window still open
> lines(x=c(25,25),y=c(1,30),lwd=2)
> lines(x=c(50,50),y=c(1,30),lwd=2)
> lines(x=c(75,75),y=c(1,30),lwd=2)
> lines(x=c(75,75),y=c(-10,30),lwd=2)
(Note that all three lines are the same height - if they seem different that is an optical illusion!) Note that we can extend the lines a little bit beyond the dimensions given (say, in this case, using: lines(x=c(50,50),y=c(-10,40),lwd=2), for instance); however we cannot extend them to the edges of the plotting window unless we use: plot.phylo(...,no.margin=TRUE). Without the margin, what we did above would look as follows:
> x<-plot(tree,no.margin=TRUE)
> lines(x=c(25,25),y=c(-10,40),lwd=2)
> lines(x=c(50,50),y=c(-10,40),lwd=2)
> lines(x=c(75,75),y=c(-10,40),lwd=2)
We can do pretty much the same thing using phytools plotTree or plotSimmap, except that we need to keep in mind that these functions automatically rescale the horizontal plotting area (including labels) to unit length. To find the total height of the rescaled tree, we need to subtract the font size × the maximum string width of the tip labels from 1. So, in the case of plotSimmap, we would do:
> # simulate a character history
> tree<-sim.history(tree,Q=matrix(c(-1/20,1/20,1/20,-1/20),2,2))
> # set colors for plotting
> cols<-c("red","blue"); names(cols)<-c(1,2)
> # plot tree
> f<-1 # font size
> plotSimmap(tree,cols,pts=F,fsize=f)
> # add lines
> h<-1-f*max(strwidth(tree$tip.label))
> lines(x=c(0.25*h,0.25*h),y=c(-10,40),lwd=2)
> lines(x=c(0.5*h,0.5*h),y=c(-10,40),lwd=2)
> lines(x=c(0.75*h,0.75*h),y=c(-10,40),lwd=2)
Fot fun, let's combine this with make.era.map:
> tree<-make.era.map(tree,c(0,25,50,75,100))
> plotSimmap(tree,lwd=3,pts=F)
> h<-1-max(strwidth(tree$tip.label))
> lines(x=c(0.25*h,0.25*h),y=c(-10,40),lwd=3,col="red")
> lines(x=c(0.5*h,0.5*h),y=c(-10,40),lwd=3,col="green")
> lines(x=c(0.75*h,0.75*h),y=c(-10,40),lwd=3,col="blue")
Pretty cool.
Wednesday, May 30, 2012
Tuesday, May 29, 2012
New version of findMRCA for large trees
I just posted a new version of findMRCA that also works for very large trees. This is accomplished by using the new function (fastMRCA) that I posted earlier today. This function still calls nodeHeights, so for extremely large trees, it is still quite slow, but I it can still compute for up to at least 60,000 tips.
Just to reminder readers, findMRCA finds the MRCA of a list of species in a vector. It originally used the ape utility function mrca - but the problem with using that function is that it computes a n × n matrix of MRCAs for n species. This will be prohibitive for larger phylogenies.
The difference between the new and old versions of this function are even apparent even for relatively small trees. Let's try a tree with just 200 tips:
> require(phytools)
> tree<-rtree(n=200)
> system.time(a<-findMRCA(tree,c("t102","t112","t38","t145")))
user system elapsed
1.48 0.00 1.50
> a
[1] 279
> source("findMRCA.R")
> system.time(b<-findMRCA(tree,c("t102","t112","t38","t145")))
user system elapsed
0.06 0.00 0.06
> b
[1] 279
Direct link to the new function is here. I have also posted a new nonstatic version of phytools with the updated function. It can be downloaded from the following link and installed from source.
Just to reminder readers, findMRCA finds the MRCA of a list of species in a vector. It originally used the ape utility function mrca - but the problem with using that function is that it computes a n × n matrix of MRCAs for n species. This will be prohibitive for larger phylogenies.
The difference between the new and old versions of this function are even apparent even for relatively small trees. Let's try a tree with just 200 tips:
> require(phytools)
> tree<-rtree(n=200)
> system.time(a<-findMRCA(tree,c("t102","t112","t38","t145")))
user system elapsed
1.48 0.00 1.50
> a
[1] 279
> source("findMRCA.R")
> system.time(b<-findMRCA(tree,c("t102","t112","t38","t145")))
user system elapsed
0.06 0.00 0.06
> b
[1] 279
Direct link to the new function is here. I have also posted a new nonstatic version of phytools with the updated function. It can be downloaded from the following link and installed from source.
Function to efficiently return the MRCA of a pair of species for trees with many tips
A user reports that she wants to use the phytools function findMRCA to get the MRCA of a group of species. The problem is that her phylogeny is very large (~60,000 tips) and findMRCA uses the ape utility function mrca which does not work for very large trees. (The reason that mrca will not work for trees this large is probably because mrca returns a giant matrix containing the MRCA of every pair of species in the tree. For a tree with 60K taxa, this matrix would require over 14GB of memory.) She asks if there might be a different way to accomplish this task.
The way I approached this problem was first just to see if there was a more efficient way to get the MRCA of a single pair of species. There is. Here, I use the function Ancestors from the phangorn package:
fastMRCA<-function(tree,sp1,sp2){
x<-match(sp1,tree$tip.label)
y<-match(sp2,tree$tip.label)
a<-Ancestors(tree,x)
b<-Ancestors(tree,y)
z<-a%in%b
return(a[min(which(z))])
}
The code x<-match(sp1,tree$tip.label) translates the input tip name to a node number; a<-Ancestors(tree,x) returns a vector containing the node numbers of the ancestors of sp1, from the tip to the root; finally z<-a%in%b and a[min(which(z))] finds the elements of a that are in b, and then identifies which of these common elements are closest to the tips of the tree (i.e., the most recent). That's it! It seems to work, even on very large trees. The next natural step is use the same approach to find the MRCA of a set of species, just as in findMRCA.
The way I approached this problem was first just to see if there was a more efficient way to get the MRCA of a single pair of species. There is. Here, I use the function Ancestors from the phangorn package:
fastMRCA<-function(tree,sp1,sp2){
x<-match(sp1,tree$tip.label)
y<-match(sp2,tree$tip.label)
a<-Ancestors(tree,x)
b<-Ancestors(tree,y)
z<-a%in%b
return(a[min(which(z))])
}
The code x<-match(sp1,tree$tip.label) translates the input tip name to a node number; a<-Ancestors(tree,x) returns a vector containing the node numbers of the ancestors of sp1, from the tip to the root; finally z<-a%in%b and a[min(which(z))] finds the elements of a that are in b, and then identifies which of these common elements are closest to the tips of the tree (i.e., the most recent). That's it! It seems to work, even on very large trees. The next natural step is use the same approach to find the MRCA of a set of species, just as in findMRCA.
Monday, May 21, 2012
New version of evolvcv.lite for multistate mapped trait
The existing phytools function evolvcv.lite fits the model of evol.vcv and Revell & Collar (2009), but with various constraints on the evolutionary rate matrices. Specifically, for two quantitative traits and a binary mapped character, evolvcv.lite fits the following four models: the same set of rates and single correlation for both states of the binary character (model 1 - this is the one rate matrix model in evol.vcv); different rates but the same correlation (model 2); different correlations but the same rates (model 3); and, finally, different correlations and rates (model 4 - this is the two rate matrix model from evol.vcv). The reason that I have not so far implemented these partially constrained models for more than two quantitative traits is because doing so constitutes a very challenging problem computationally. However there is no reason why the function could not be expanded to a mapped multistate character (rather than simply a binary character, as in the present implementation). In fact, a phytools user and blog reader recently requested this addition and I have finally gotten around to implementing and debugging this.
This version also fixes a bug in the previous edition which miscalculated the number of parameters (by -1 in all cases), and thus the AIC scores as well.
The new version of this function is here. I have also built a new version of phytools (here) which can be downloaded and installed from source.
One neat trick that I learned in the process of programming this update was how to add corresponding elements of list of matrices. Say, for instance, that X is a list of matrices of identical dimensions. It turns out that sumX<-Reduce("+",X) gives the sum of the matrices in X. Who knew!
Let's try out the new version of evolvcv.lite with an example.
> # install new version of phytools
> install.packages("phytools_0.1-82.tar.gz",type="source", repos=NULL)
Installing package(s) into ‘C:/R/win-library/2.14’
...
* DONE (phytools)
> require(phytools)
Loading required package: phytools
...
> # simulate tree & discrete character history
> tree<-sim.history(pbtree(n=100,scale=1), Q=matrix(c(-2,1,1,1,-2,1,1,1,-2),3,3))
> # simulate uncorrelated traits with three different rates
> # depending on the state
> X<-cbind(sim.rates(tree,c(1,2,3)),sim.rates(tree,c(1,2,3)))
> # fit models
> R<-evolvcv.lite(tree,X)
> # in the following I have hidden some results for brevity
> # only the full output for the best fitting model is shown
> R
$model1
$model1$description
[1] "common rates, common correlation"
...
$model1$AIC
[1] 379.6677
$model2
$model2$description
[1] "different rates, common correlation"
$model2$R
$model2$R$`1`
[,1] [,2]
[1,] 1.218973 0.096771
[2,] 0.096771 1.244826
$model2$R$`2`
[,1] [,2]
[1,] 1.6109460 0.1451599
[2,] 0.1451599 2.1194570
$model2$R$`3`
[,1] [,2]
[1,] 1.6255930 0.2015308
[2,] 0.2015308 4.0483984
$model2$logLik
[1] -181.8455
$model2$convergence
[1] 0
$model2$k
[1] 9
$model2$AIC
[1] 381.6911
$model3
$model3$description
[1] "common rates, different correlation"
...
$model3$AIC
[1] 382.892
$model4
$model4$description
[1] "no common structure"
...
$model4$AIC
[1] 385.1049
Cool. The best fitting model is our generating model in this case: common correlation but different rates depending on the state of our multistate character.
This version also fixes a bug in the previous edition which miscalculated the number of parameters (by -1 in all cases), and thus the AIC scores as well.
The new version of this function is here. I have also built a new version of phytools (here) which can be downloaded and installed from source.
One neat trick that I learned in the process of programming this update was how to add corresponding elements of list of matrices. Say, for instance, that X is a list of matrices of identical dimensions. It turns out that sumX<-Reduce("+",X) gives the sum of the matrices in X. Who knew!
Let's try out the new version of evolvcv.lite with an example.
> # install new version of phytools
> install.packages("phytools_0.1-82.tar.gz",type="source", repos=NULL)
Installing package(s) into ‘C:/R/win-library/2.14’
...
* DONE (phytools)
> require(phytools)
Loading required package: phytools
...
> # simulate tree & discrete character history
> tree<-sim.history(pbtree(n=100,scale=1), Q=matrix(c(-2,1,1,1,-2,1,1,1,-2),3,3))
> # simulate uncorrelated traits with three different rates
> # depending on the state
> X<-cbind(sim.rates(tree,c(1,2,3)),sim.rates(tree,c(1,2,3)))
> # fit models
> R<-evolvcv.lite(tree,X)
> # in the following I have hidden some results for brevity
> # only the full output for the best fitting model is shown
> R
$model1
$model1$description
[1] "common rates, common correlation"
...
$model1$AIC
[1] 379.6677
$model2
$model2$description
[1] "different rates, common correlation"
$model2$R
$model2$R$`1`
[,1] [,2]
[1,] 1.218973 0.096771
[2,] 0.096771 1.244826
$model2$R$`2`
[,1] [,2]
[1,] 1.6109460 0.1451599
[2,] 0.1451599 2.1194570
$model2$R$`3`
[,1] [,2]
[1,] 1.6255930 0.2015308
[2,] 0.2015308 4.0483984
$model2$logLik
[1] -181.8455
$model2$convergence
[1] 0
$model2$k
[1] 9
$model2$AIC
[1] 381.6911
$model3
$model3$description
[1] "common rates, different correlation"
...
$model3$AIC
[1] 382.892
$model4
$model4$description
[1] "no common structure"
...
$model4$AIC
[1] 385.1049
Cool. The best fitting model is our generating model in this case: common correlation but different rates depending on the state of our multistate character.
Tuesday, May 15, 2012
Addendum to previous post on plotting slanted phylograms with phytools::phenogram
In my last post I demonstrated how the phytools function phenogram could be used to plot a slanted phylogram - at least most of the time. The ape phylogenetics package function plot.phylo can also plot slanted phylograms (by setting type="cladogram" for a tree with branch lengths assigned), but this phylogram has a fundamentally different style than the type created by phenogram. The effect in particular is one in which the branches of the tree plotted with phenogram slant much more gently from the root than the branches in plot.phylo(...,type="cladogram"). The following graphic contrasts the two styles:
While I was musing about yesterday's post this morning, it occurred to me that plot.phylo(...,type="cladogram") kind of looks like it uses the same method as phenogram, but with the positions of the internal nodes determined by computing the "ancestral state" (that is, of the y coordinate of the tip) for the root node of each subtree separately. This could be accomplished using ace(...,method="pic") in the ape package. Well, this is not, in fact, how plot.phylo(...,type="cladogram") works, but the two methods visually present as almost identical. For instance:
> set.seed(1)
> tree<-read.tree(text=write.tree(pbtree(n=50)))
> plot.phylo(tree,type="cladogram")
> x<-1:50; names(x)<-tree$tip.label
> a<-ace(x,tree,method="pic")$ace
> x11(); phenogram(tree,c(x,a))
It turns out that both of these methods can sometimes result in lines that cross. For instance:
> set.seed(25)
> tree<-pbtree(n=50)
> plot.phylo(tree,type="cladogram")
Now, I'm not sure whether to characterize this as a bug in plot.phylo or not. Evidently, it is not always possible to avoid having lines cross when we want our tip labels to be evenly spaced on the vertical axis. This is stated in Felsenstein (2008; p. 576): "Note that with many of these methods (for plotting slanted phylograms), one cannot always avoid having lines cross. . . . The only way of avoiding crossing is to have the tips not be evenly spaced along the y axis." Clearly, in the above plotted example it would be possible to redraw this tree with the same horizontal branch lengths and no crossing lines, no?
While I was musing about yesterday's post this morning, it occurred to me that plot.phylo(...,type="cladogram") kind of looks like it uses the same method as phenogram, but with the positions of the internal nodes determined by computing the "ancestral state" (that is, of the y coordinate of the tip) for the root node of each subtree separately. This could be accomplished using ace(...,method="pic") in the ape package. Well, this is not, in fact, how plot.phylo(...,type="cladogram") works, but the two methods visually present as almost identical. For instance:
> set.seed(1)
> tree<-read.tree(text=write.tree(pbtree(n=50)))
> plot.phylo(tree,type="cladogram")
> x<-1:50; names(x)<-tree$tip.label
> a<-ace(x,tree,method="pic")$ace
> x11(); phenogram(tree,c(x,a))
It turns out that both of these methods can sometimes result in lines that cross. For instance:
> set.seed(25)
> tree<-pbtree(n=50)
> plot.phylo(tree,type="cladogram")
Now, I'm not sure whether to characterize this as a bug in plot.phylo or not. Evidently, it is not always possible to avoid having lines cross when we want our tip labels to be evenly spaced on the vertical axis. This is stated in Felsenstein (2008; p. 576): "Note that with many of these methods (for plotting slanted phylograms), one cannot always avoid having lines cross. . . . The only way of avoiding crossing is to have the tips not be evenly spaced along the y axis." Clearly, in the above plotted example it would be possible to redraw this tree with the same horizontal branch lengths and no crossing lines, no?
Monday, May 14, 2012
Neat way to plot a slanted phylogram in phytools that almost always works
I'm not sure what I was doing when I thought to try this, but I realized that the phytools function phenogram can also be used to plot a neat looking slanted phylogram. This works as follows:
> tree<-pbtree(n=20,scale=100)
> # we do the following to make sure the tips are
> # in the right order
> tree<-read.tree(text=write.tree(tree))
> x<-1:length(tree$tip)
> names(x)<-tree$tip.label
> phenogram(tree,x)
Here the horizontal axis is time since the root, and the vertical axis doesn't mean anything of course. Unfortunately, as alluded in the post title, this only works most of the time - particularly when the tree has a lot of tips. For example, for one stochastic 50 taxon tree I get the following problem:
Obviously, the branches of the tree cross in a couple of different spots, which is no good. If we are determined to plot a tree in this format, we could try rotating the offending nodes above the problematic crossings and then replot our tree:
> plotTree(tree,node.numbers=T)
> tree<-rotate(tree,95)
> tree<-rotate(tree,53)
> tree<-read.tree(text=write.tree(tree))
> x<-1:length(tree$tip)
> names(x)<-tree$tip.label
> phenogram(tree,x)
It worked - at least for this particular case.
Some users might note that ape can also plot a slanted phylogram:
> plot.phylo(tree,type="cladogram")
But, obviously, this visualization has a very different look compared to the one we created previously. I'm not aware of other packages that might have an analogous function. Perhaps some readers of this blog are.
> tree<-pbtree(n=20,scale=100)
> # we do the following to make sure the tips are
> # in the right order
> tree<-read.tree(text=write.tree(tree))
> x<-1:length(tree$tip)
> names(x)<-tree$tip.label
> phenogram(tree,x)
Here the horizontal axis is time since the root, and the vertical axis doesn't mean anything of course. Unfortunately, as alluded in the post title, this only works most of the time - particularly when the tree has a lot of tips. For example, for one stochastic 50 taxon tree I get the following problem:
Obviously, the branches of the tree cross in a couple of different spots, which is no good. If we are determined to plot a tree in this format, we could try rotating the offending nodes above the problematic crossings and then replot our tree:
> plotTree(tree,node.numbers=T)
> tree<-rotate(tree,95)
> tree<-rotate(tree,53)
> tree<-read.tree(text=write.tree(tree))
> x<-1:length(tree$tip)
> names(x)<-tree$tip.label
> phenogram(tree,x)
It worked - at least for this particular case.
Some users might note that ape can also plot a slanted phylogram:
> plot.phylo(tree,type="cladogram")
But, obviously, this visualization has a very different look compared to the one we created previously. I'm not aware of other packages that might have an analogous function. Perhaps some readers of this blog are.
Friday, May 11, 2012
Painting different clades with different colors in phenogram function
A user recently submitted the following question:
is it possible to create a phenogram (that is, a projection of the tree into a two dimensional space defined by morphology on the vertical axis and time, see here) with different clades as different colors?
The function phenogram already allows a mapped discrete character, so if we want to color arbitrary clades on our tree different colors, we just need to combine phenogram with the phytools function paintSubTree, described here.
Let's try it in the following illustrating example:
> # first simulate a random tree
> tree<-pbtree(n=20,scale=100)
> # now plot the tree with node labels
> # so we can select the clades we want to paint
> plotTree(tree,pts=F,node.numbers=T)
OK, this tree has two main clades: one descended from node number "22", and the other descended from node number "26". Let's first paint the former clade with state "2" and the latter clade with state "3", we can leave the stem branches of each clade in state "1" (although we need not).
> tree<-paintSubTree(tree,node=22,state="2")
> tree<-paintSubTree(tree,node=26,state="3")
> # now let's plot using plotSimmap to ensure
> # that the correct branches were painted
> cols<-c("black","blue","red"); names(cols)<-1:3
> plotSimmap(tree,cols,pts=F,lwd=3,node.numbers=T)
Finally, let's simulate some data (normally, of course, we would read our data from file) and plot a traitgram using the phenogram function in phytools:
> x<-fastBM(tree)
> phenogram(tree,x,colors=cols,fsize=0.8)
That's it.
is it possible to create a phenogram (that is, a projection of the tree into a two dimensional space defined by morphology on the vertical axis and time, see here) with different clades as different colors?
The function phenogram already allows a mapped discrete character, so if we want to color arbitrary clades on our tree different colors, we just need to combine phenogram with the phytools function paintSubTree, described here.
Let's try it in the following illustrating example:
> # first simulate a random tree
> tree<-pbtree(n=20,scale=100)
> # now plot the tree with node labels
> # so we can select the clades we want to paint
> plotTree(tree,pts=F,node.numbers=T)
OK, this tree has two main clades: one descended from node number "22", and the other descended from node number "26". Let's first paint the former clade with state "2" and the latter clade with state "3", we can leave the stem branches of each clade in state "1" (although we need not).
> tree<-paintSubTree(tree,node=22,state="2")
> tree<-paintSubTree(tree,node=26,state="3")
> # now let's plot using plotSimmap to ensure
> # that the correct branches were painted
> cols<-c("black","blue","red"); names(cols)<-1:3
> plotSimmap(tree,cols,pts=F,lwd=3,node.numbers=T)
Finally, let's simulate some data (normally, of course, we would read our data from file) and plot a traitgram using the phenogram function in phytools:
> x<-fastBM(tree)
> phenogram(tree,x,colors=cols,fsize=0.8)
That's it.
Thursday, May 10, 2012
New version of drop.tip.simmap
OK, I just posted a new version of drop.tip.simmap which modifies both the $maps and $mapped.edge elements of the modified tree object created by (say) read.simmap or sim.history. Direct link to the code is here, but I have also built a new version (0.1-81) of phytools containing this update. You can download and install from source here.
Nothing spectacular, I just created this function from the tree pruning function that I completed (with some errors, now fixed in this version) this afternoon. Let's check it out:
> # first load from source or install new phytools
> install.packages("phytools_0.1-81.tar.gz",type="source", repos=NULL)
* installing *source* package 'phytools' ...
...
* DONE (phytools)
> require(phytools)
Loading required package: phytools
...
> # now simulate tree or read from file (here simulated)
> tree<-pbtree(n=20,scale=2)
> Q<-matrix(c(-2,1,1,1,-2,1,1,1,-2),3,3)
> mtree<-sim.history(tree,Q)
> # this is the full, unpruned tree
> plotSimmap(mtree,pts=F,lwd=3)
> # pick some tips to prune at random
> tips<-sample(tree$tip.label,5)
> tips
[1] "t20" "t8" "t5" "t16" "t14"
> ptree<-drop.tip.simmap(mtree,tips)
> plotSimmap(ptree,lwd=3,pts=F)
Well, this seems to work - but I welcome feedback from users that try it on their own data or from trees read from file. Good luck!
Nothing spectacular, I just created this function from the tree pruning function that I completed (with some errors, now fixed in this version) this afternoon. Let's check it out:
> # first load from source or install new phytools
> install.packages("phytools_0.1-81.tar.gz",type="source", repos=NULL)
* installing *source* package 'phytools' ...
...
* DONE (phytools)
> require(phytools)
Loading required package: phytools
...
> # now simulate tree or read from file (here simulated)
> tree<-pbtree(n=20,scale=2)
> Q<-matrix(c(-2,1,1,1,-2,1,1,1,-2),3,3)
> mtree<-sim.history(tree,Q)
> # this is the full, unpruned tree
> plotSimmap(mtree,pts=F,lwd=3)
> # pick some tips to prune at random
> tips<-sample(tree$tip.label,5)
> tips
[1] "t20" "t8" "t5" "t16" "t14"
> ptree<-drop.tip.simmap(mtree,tips)
> plotSimmap(ptree,lwd=3,pts=F)
Well, this seems to work - but I welcome feedback from users that try it on their own data or from trees read from file. Good luck!
"Mini" version of drop.tip
The phytools package can read, simulate, plot, reorder, and write stochastic character map style trees. (That is, trees in which a discrete character history is stored on the edges of the tree.) I even recently added the capacity to read SIMMAP v1.5 trees to the function read.simmap. Unfortunately, phytools only has a very limited capacity to manipulate SIMMAP style trees. In particular, the one existing function (drop.tip.simmap only alters the $mapped.edge element of the modified "phylo" object, without touching $maps. $mapped.edge contains only the time spent in each state on each edge, while $maps contains the times and order of each state on each edge. The latter element is required for both plotting (using plotSimmap) and for other types of analyses using (for instance) the new OUwie phylogenetics package.
Well, I'm at long last trying to build a function that will drop tips from the tree, but preserve both $maps and $mapped.edge. At first, I was hoping to do this using basically the same trick (but in reverse) that I used to read SIMMAP trees from file in the first version of read.simmap (this trick has long been replaced by a much more sophisticated function). Alas, this did not work. So I have gone back to the basics and I have started out by writing a basic tip dropping function of my own. This is incredibly simple, and much less versatile than the ape function drop.tip, but it gives me a structure to work from to build up a full drop.tip.simmap function.
The function is as follows:
pruneTree<-function(tree,tip){
tip<-which(tree$tip.label%in%tip)
edges<-match(tip,tree$edge[,2])
tree$edge<-tree$edge[setdiff(1:nrow(tree$edge),edges),]
z<-setdiff(tree$edge[,2],tree$edge[,1])
z<-z[z>length(tree$tip)]
while(length(z)>0){
edges<-match(z,tree$edge[,2])
tree$edge<-tree$edge[setdiff(1:nrow(tree$edge),edges),]
z<-setdiff(tree$edge[,2],tree$edge[,1])
z<-z[z>length(tree$tip)]
}
z<-setdiff(tree$edge[,2],tree$edge[,1])
tree$tip.label<-tree$tip.label[z]
tree$edge[which(tree$edge[,2]%in%z),2]<-1:length(tree$tip)
i<-1
while(i < nrow(tree$edge)){
single<-sum(tree$edge[i,2]==tree$edge[,1])==1
while(single){
if(sum(tree$edge[i,2]==tree$edge[,1])==1){
z<-match(tree$edge[i,2],tree$edge[,1])
tree$edge[i,2]<-tree$edge[z,2]
tree$edge<-tree$edge[setdiff(1:nrow(tree$edge),z),]
}
single<-sum(tree$edge[i,2]==tree$edge[,1])==1
}
i<-i+1
}
z<-unique(as.vector(tree$edge))
z<-z[z>length(tree$tip)]
y<-order(z)+length(tree$tip)
for(i in 1:length(z)) tree$edge[tree$edge==z[i]]<-y[i]
tree$Nnode<-max(tree$edge)-length(tree$tip)
tree$edge.length<-NULL
class(tree)<-"phylo"
return(tree)
}
OK, let's try it:
> require(phytools); source("pruneTree.R")
> set.seed(1)
> tree<-pbtree(n=20); tree$edge.length<-NULL
> plot(tree)
> ptree<-pruneTree(tree,c("t6","t8","t7","t3","t2","t15"))
> plot(ptree)
Now I just need to figure out how to preserve the mappings and I'll be all set!
Well, I'm at long last trying to build a function that will drop tips from the tree, but preserve both $maps and $mapped.edge. At first, I was hoping to do this using basically the same trick (but in reverse) that I used to read SIMMAP trees from file in the first version of read.simmap (this trick has long been replaced by a much more sophisticated function). Alas, this did not work. So I have gone back to the basics and I have started out by writing a basic tip dropping function of my own. This is incredibly simple, and much less versatile than the ape function drop.tip, but it gives me a structure to work from to build up a full drop.tip.simmap function.
The function is as follows:
pruneTree<-function(tree,tip){
tip<-which(tree$tip.label%in%tip)
edges<-match(tip,tree$edge[,2])
tree$edge<-tree$edge[setdiff(1:nrow(tree$edge),edges),]
z<-setdiff(tree$edge[,2],tree$edge[,1])
z<-z[z>length(tree$tip)]
while(length(z)>0){
edges<-match(z,tree$edge[,2])
tree$edge<-tree$edge[setdiff(1:nrow(tree$edge),edges),]
z<-setdiff(tree$edge[,2],tree$edge[,1])
z<-z[z>length(tree$tip)]
}
z<-setdiff(tree$edge[,2],tree$edge[,1])
tree$tip.label<-tree$tip.label[z]
tree$edge[which(tree$edge[,2]%in%z),2]<-1:length(tree$tip)
i<-1
while(i < nrow(tree$edge)){
single<-sum(tree$edge[i,2]==tree$edge[,1])==1
while(single){
if(sum(tree$edge[i,2]==tree$edge[,1])==1){
z<-match(tree$edge[i,2],tree$edge[,1])
tree$edge[i,2]<-tree$edge[z,2]
tree$edge<-tree$edge[setdiff(1:nrow(tree$edge),z),]
}
single<-sum(tree$edge[i,2]==tree$edge[,1])==1
}
i<-i+1
}
z<-unique(as.vector(tree$edge))
z<-z[z>length(tree$tip)]
y<-order(z)+length(tree$tip)
for(i in 1:length(z)) tree$edge[tree$edge==z[i]]<-y[i]
tree$Nnode<-max(tree$edge)-length(tree$tip)
tree$edge.length<-NULL
class(tree)<-"phylo"
return(tree)
}
OK, let's try it:
> require(phytools); source("pruneTree.R")
> set.seed(1)
> tree<-pbtree(n=20); tree$edge.length<-NULL
> plot(tree)
> ptree<-pruneTree(tree,c("t6","t8","t7","t3","t2","t15"))
> plot(ptree)
Now I just need to figure out how to preserve the mappings and I'll be all set!
Tuesday, May 8, 2012
Getting the ancestral state estimate from the "same" node on many trees
An R-SIG-phylo user today asked how to extract the ancestral state estimate from the same node on a set of trees. First, we need to define what is meant by the "same" node. Here, I presume that the same node is defined by the set of taxa descended from the that node (so is equivalently the MRCA of a set of tip species). In a set of trees read into R using read.tree there is no guarantee that a node defined in this way will have the same node number in every tree.
I suggested that one could use the phytools function findMRCA, which returns the most recent common ancestor for a set of tip species provided in a vector. Say our set of tip species consists of "t1", "t3", and "t8", to extract the estimated ancestral value for their MRCA on a set of trees, we can just use the following code (trait vector is in x):
> sp<-c("t1","t3","t8")
> target.state<-sapply(trees,function(a,x,sp) ace(x,a)$ace[as.character(findMRCA(a,sp))],x=x,sp=sp)
Simple as that. Of course, we can also might want to check in each case as to whether our tip species represent a monophyletic group. To do that we could just do:
> m<-sapply(trees,is.monophyletic,tips=sp)
And then to pull out only the target node state estimates from trees in which our list of taxa are monophyletic, you can use:
> target.state<-target.state[m]
That's it.
I suggested that one could use the phytools function findMRCA, which returns the most recent common ancestor for a set of tip species provided in a vector. Say our set of tip species consists of "t1", "t3", and "t8", to extract the estimated ancestral value for their MRCA on a set of trees, we can just use the following code (trait vector is in x):
> sp<-c("t1","t3","t8")
> target.state<-sapply(trees,function(a,x,sp) ace(x,a)$ace[as.character(findMRCA(a,sp))],x=x,sp=sp)
Simple as that. Of course, we can also might want to check in each case as to whether our tip species represent a monophyletic group. To do that we could just do:
> m<-sapply(trees,is.monophyletic,tips=sp)
And then to pull out only the target node state estimates from trees in which our list of taxa are monophyletic, you can use:
> target.state<-target.state[m]
That's it.
Friday, May 4, 2012
Ordering trees in a multiPhylo object by the number of tips
Today a R phylogenetics user asks if there is an easy way to sort the trees in a "multiPhylo" object by the number of tips each tree contains. The answer is yes. One way to do it is as follows:
> # first simulate a set of trees with different numbers of tips
> require(phytools)
> trees<-replicate(10,pbtree(n=round(runif(n=1,min=10,max=40))), simplify=FALSE)
> class(trees)<-"multiPhylo"
> trees
10 phylogenetic trees
> # now get the number of tips in each tree
> ntips<-sapply(trees,function(x) length(x$tip))
> ntips
[1] 35 19 13 34 18 27 18 33 10 25
> # now sort by the decreasing order of ntips
> sorted.trees<-trees[order(ntips,decreasing=T)]
To check & see that we have sorted our trees we can of course just plot them using plot.multiPhylo, but we can also just recalculate the vector ntips from our new list of trees:
> ntips.sorted<-sapply(sorted.trees,function(x) length(x$tip))
> ntips.sorted
[1] 35 34 33 27 25 19 18 18 13 10
Cool - sorted!
The same user also asked how to pull NULL trees from the list. I suggested one could just do the following:
> trees.no.null<-lapply(trees,function(x) if(!is.null(x)) x))
> class(trees.no.null)<-"multiPhylo"
Evidently, this works - but it just occurred to me that the following simpler option should also do the trick:
> trees.no.null<-trees[!is.null(trees)]
Cool.
> # first simulate a set of trees with different numbers of tips
> require(phytools)
> trees<-replicate(10,pbtree(n=round(runif(n=1,min=10,max=40))), simplify=FALSE)
> class(trees)<-"multiPhylo"
> trees
10 phylogenetic trees
> # now get the number of tips in each tree
> ntips<-sapply(trees,function(x) length(x$tip))
> ntips
[1] 35 19 13 34 18 27 18 33 10 25
> # now sort by the decreasing order of ntips
> sorted.trees<-trees[order(ntips,decreasing=T)]
To check & see that we have sorted our trees we can of course just plot them using plot.multiPhylo, but we can also just recalculate the vector ntips from our new list of trees:
> ntips.sorted<-sapply(sorted.trees,function(x) length(x$tip))
> ntips.sorted
[1] 35 34 33 27 25 19 18 18 13 10
Cool - sorted!
The same user also asked how to pull NULL trees from the list. I suggested one could just do the following:
> trees.no.null<-lapply(trees,function(x) if(!is.null(x)) x))
> class(trees.no.null)<-"multiPhylo"
Evidently, this works - but it just occurred to me that the following simpler option should also do the trick:
> trees.no.null<-trees[!is.null(trees)]
Cool.
Thursday, May 3, 2012
Specifying node colors with phylomorphospace()
A user recently made the following request:
I was just wondering if you would mind posting a short example of the code and data format used to specify colours in the phylomorphospace function (e.g., to create the figure form the paper describing the packages)?
Ok, just for reference, a screen grab of that figure is here:
What this figure shows is a projection of the phylogeny of Caribbean Anolis ecomorph species into a two dimensional morphospace defined by PC 1 & PC 2 from the morphological dataset and tree of Mahler et al. (2010). The tip nodes are colored by ecomorph category; and the internal nodes are black.
All right, for simplicity I'll assume that we want to replicate this general plot in a hypothetical case. First, let's simulate a tree and data for two characters:
> require(phytools)
> tree<-pbtree(n=21)
> X<-cbind(fastBM(tree),fastBM(tree))
Here, I have just simulated the tree & data; normally we would probably read these from file using functions like read.tree or read.nexus and read.csv. Here is a visualization of our simulated tree:
> plotTree(tree,pts=F)
Next, let's say we want to color (for simplicity) every third tip red, blue, or green. (I.e., the series t1, t4, t7, etc. is red.) Normally, we would probably read this from a file as well. For instance, in the ecomorph example, above, we would read a data file containing the ecomorph identification of each of the species from the tree. We could then settle on a color scheme for translation and convert our ecomorph codes to colors. In our example here, we can just do:
> tip.cols<-rep(c("red","blue","green"),7)
> names(tip.cols)<-tree$tip.label
> cols<-c(tip.cols[tree$tip.label],rep("black",tree$Nnode))
> names(cols)<-1:(length(tree$tip)+tree$Nnode)
> phylomorphospace(tree,X,control=list(col.node=cols))
If you examine this figure closely, you'll see that (as intended) everything third numbered tip node is colored red, blue, and green. Obviously, with a real (rather than simulated) tree and dataset we would read in the states for the tip nodes from a file (and they would most likely mean something). It is important to both reorder the color vector by tree$tip.label and re-number it by node number. Don't forget to add elements for internal nodes, even if they are to be colored black (for now at least: perhaps I will fix this in a future version). If we just want to color tip nodes, this is not difficult because the node numbers of the tips are 1 through N (for N species) in the order given by tree$tip.label. It is also probably a good idea to provide X as a matrix & control$col.node as a named vector, just to be safe.
I hope this helps!
I was just wondering if you would mind posting a short example of the code and data format used to specify colours in the phylomorphospace function (e.g., to create the figure form the paper describing the packages)?
Ok, just for reference, a screen grab of that figure is here:
What this figure shows is a projection of the phylogeny of Caribbean Anolis ecomorph species into a two dimensional morphospace defined by PC 1 & PC 2 from the morphological dataset and tree of Mahler et al. (2010). The tip nodes are colored by ecomorph category; and the internal nodes are black.
All right, for simplicity I'll assume that we want to replicate this general plot in a hypothetical case. First, let's simulate a tree and data for two characters:
> require(phytools)
> tree<-pbtree(n=21)
> X<-cbind(fastBM(tree),fastBM(tree))
Here, I have just simulated the tree & data; normally we would probably read these from file using functions like read.tree or read.nexus and read.csv. Here is a visualization of our simulated tree:
> plotTree(tree,pts=F)
Next, let's say we want to color (for simplicity) every third tip red, blue, or green. (I.e., the series t1, t4, t7, etc. is red.) Normally, we would probably read this from a file as well. For instance, in the ecomorph example, above, we would read a data file containing the ecomorph identification of each of the species from the tree. We could then settle on a color scheme for translation and convert our ecomorph codes to colors. In our example here, we can just do:
> tip.cols<-rep(c("red","blue","green"),7)
> names(tip.cols)<-tree$tip.label
> cols<-c(tip.cols[tree$tip.label],rep("black",tree$Nnode))
> names(cols)<-1:(length(tree$tip)+tree$Nnode)
> phylomorphospace(tree,X,control=list(col.node=cols))
If you examine this figure closely, you'll see that (as intended) everything third numbered tip node is colored red, blue, and green. Obviously, with a real (rather than simulated) tree and dataset we would read in the states for the tip nodes from a file (and they would most likely mean something). It is important to both reorder the color vector by tree$tip.label and re-number it by node number. Don't forget to add elements for internal nodes, even if they are to be colored black (for now at least: perhaps I will fix this in a future version). If we just want to color tip nodes, this is not difficult because the node numbers of the tips are 1 through N (for N species) in the order given by tree$tip.label. It is also probably a good idea to provide X as a matrix & control$col.node as a named vector, just to be safe.
I hope this helps!
Wednesday, May 2, 2012
New version of phytools (v 0.1-8) on CRAN
The newest version of phytools (0.1-8) is now available on CRAN. Check out the phytools CRAN page here. It usually takes a couple of days for a new package version to percolate through all the CRAN mirrors.
Updates in this version (over the previous CRAN release) include:
1. A new version of read.simmap that can read SIMMAP version 1.5 tree files.
2. A new version of ltt to simultaneously create lineage-through-time plots (and compute γ) for multiple trees.
3. A bug fix in fitBayes.
4. A new version of treeSlice that returns trivial as well as non-trivial subtrees.
5. New functions getExtant and getExtinct which return a list of the tip names of all the extant species (or its complement).
Enjoy!
Updates in this version (over the previous CRAN release) include:
1. A new version of read.simmap that can read SIMMAP version 1.5 tree files.
2. A new version of ltt to simultaneously create lineage-through-time plots (and compute γ) for multiple trees.
3. A bug fix in fitBayes.
4. A new version of treeSlice that returns trivial as well as non-trivial subtrees.
5. New functions getExtant and getExtinct which return a list of the tip names of all the extant species (or its complement).
Enjoy!
Tuesday, May 1, 2012
read.simmap now reads SIMMAP v1.5 input files
I finally got around to adding the capacity to read SIMMAP v1.5 (program description here). SIMMAP v1.5 mapped tree files use a very different modified Nexus format tree file than the previous version. Although I obviously can't speak directly to the motives of the program author, my guess is that this change was designed to allow SIMMAP tree files to be read into other programs that read Nexus format trees (albeit without the mapping). This was accomplished by bracketing all mappings with [ square brackets ] which are ignored as "comments" by Nexus convention. In particular, the following Newick string from version 1.0 SIMMAP tree:
((1:{0,0.3:1,0.4:0,0.3},2:{0,1.0}):{0,1.0},(3:{1,0.75:0,0.25},4:{0,1.0}):{0,1.0});
Representing the following tree:
Would be written as follows by SIMMAP v1.5:
((1:[&map={0,0.3,1,0.4,0}]1.0,2:[&map={0}]1.0):[&map={0}]1.0,(3:[&map={1,0.75,0}]1.0,4:[&map={0}]1.0):[&map={0}]1.0);
In SIMMAP v1.5 output files the mapping information for each branch is contained in the [&maps={...}] elements in which ... contains the state and (if there is more than one state on a branch) the time spend in each state from the tipward to the most rootward states on the edge - but excluding the time spend in the most rootward state (which can here be inferred by subtracting the time spend in each tipward state from the total length of the edge given after the ]).
Version is specified by a new argument: read.simmap(...,version), which is a numeric argument and can be 1.0 or 1.5.
Like prior versions of read.simmap, this present version can also read multiple trees in a list. However, I should note that if version=1.5 the tree or trees must be supplied in a Nexus style input file (not in a text string or Phylip style file, as is allowed for version=1.0).
Ok, well I have posted the function online here. It is also part of the latest version of phytools, version 0.1-8, which can be installed from the source code from my website (phytools page) and has also been submitted to CRAN.
I have only tested this with a few input files, so if you are able to give it a go - please let me know of the result.
((1:{0,0.3:1,0.4:0,0.3},2:{0,1.0}):{0,1.0},(3:{1,0.75:0,0.25},4:{0,1.0}):{0,1.0});
Representing the following tree:
Would be written as follows by SIMMAP v1.5:
((1:[&map={0,0.3,1,0.4,0}]1.0,2:[&map={0}]1.0):[&map={0}]1.0,(3:[&map={1,0.75,0}]1.0,4:[&map={0}]1.0):[&map={0}]1.0);
In SIMMAP v1.5 output files the mapping information for each branch is contained in the [&maps={...}] elements in which ... contains the state and (if there is more than one state on a branch) the time spend in each state from the tipward to the most rootward states on the edge - but excluding the time spend in the most rootward state (which can here be inferred by subtracting the time spend in each tipward state from the total length of the edge given after the ]).
Version is specified by a new argument: read.simmap(...,version), which is a numeric argument and can be 1.0 or 1.5.
Like prior versions of read.simmap, this present version can also read multiple trees in a list. However, I should note that if version=1.5 the tree or trees must be supplied in a Nexus style input file (not in a text string or Phylip style file, as is allowed for version=1.0).
Ok, well I have posted the function online here. It is also part of the latest version of phytools, version 0.1-8, which can be installed from the source code from my website (phytools page) and has also been submitted to CRAN.
I have only tested this with a few input files, so if you are able to give it a go - please let me know of the result.
Subscribe to:
Posts (Atom)