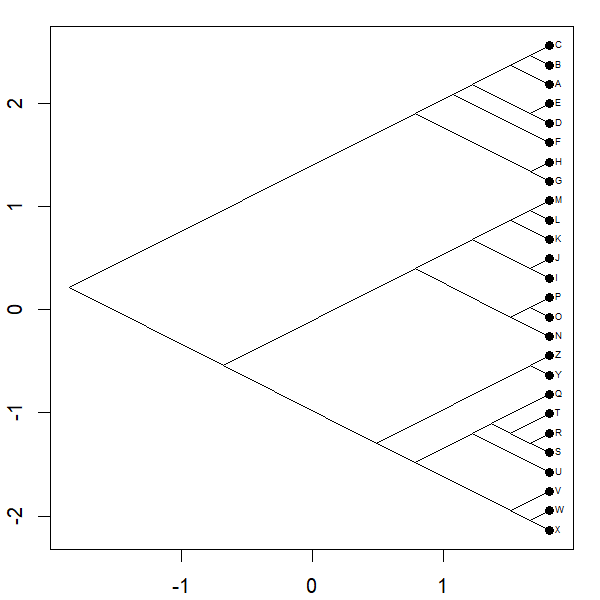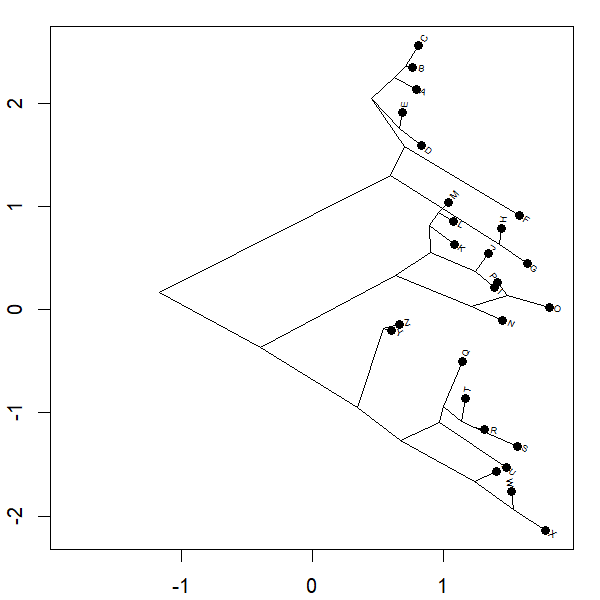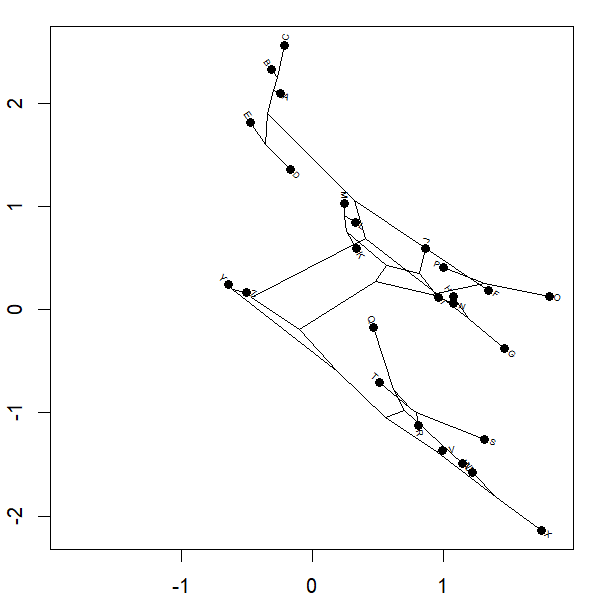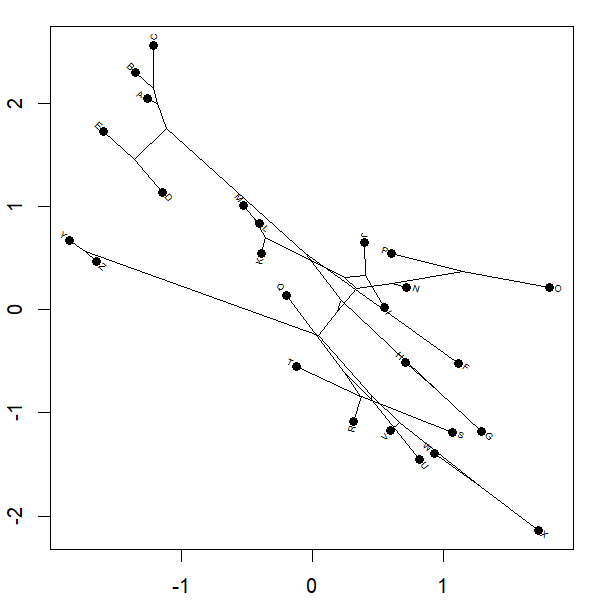
Yesterday I posted a function to animation the projection of a phylogeny into so-called 'phylomorphospace.' Basically the function morphs a cladogram into morphospace.
I have now updated this function & added it to the phytools package so it can be obtained merly by updating phytools from GitHub using devtools
The update consisted of doing a few things:
1) I eliminated a (hard to notice except when converted to .gif) first blank
plotting device using par(new=TRUE).
2) I allow users to morph backwards from phylomorphospace to tree, forwards, or both.
Complete code of the function can be seen above; however for the plot
below I also made a small 'fix' (using fix) the internally
used phylomorphospace which I intend to add as an option
later.
Here's a demo:
library(phytools)
packageVersion("phytools")
## [1] '0.6.58'
tree
##
## Phylogenetic tree with 40 tips and 39 internal nodes.
##
## Tip labels:
## t19, t20, t5, t15, t16, t8, ...
##
## Rooted; includes branch lengths.
X
## [,1] [,2]
## t19 3.50708037 -1.1096582
## t20 4.27937896 -1.2525421
## t5 0.08209894 -2.6875071
## t15 1.47192856 -3.3443560
## t16 1.74646002 -2.1723942
## t8 1.47101027 -3.9437620
## t39 3.03199098 -3.7106981
## t40 2.80301669 -4.0854970
## t13 1.95146677 -2.9904940
## t12 1.33706669 -4.8753683
## t17 0.67514002 -4.3273505
## t18 0.63260813 -2.9261122
## t2 -0.70893947 -5.9488395
## t3 -1.54193957 -4.8277646
## t6 -1.63946177 -7.8730380
## t7 -0.94456462 -8.8354219
## t9 -0.16164120 -0.6339037
## t10 2.18203703 1.4402154
## t26 0.98913114 2.3696318
## t31 1.98265867 1.3857154
## t32 1.45834021 1.7523258
## t1 0.90273795 -0.6776714
## t24 0.09296010 -0.9788181
## t25 -0.60155598 -1.0161882
## t28 0.36367701 -1.6175121
## t37 -0.21208935 -2.9695885
## t38 -0.24609233 -3.0210521
## t4 2.96819592 -1.9776346
## t23 1.77672273 -1.5263565
## t29 2.24431018 -0.4320801
## t30 2.41836111 -0.9098723
## t11 0.94537923 -1.8870267
## t27 0.20277275 0.5987702
## t33 0.66893070 -0.3095932
## t34 0.69709488 0.5173531
## t14 0.18474066 -1.4782937
## t35 2.09494024 -1.3206123
## t36 1.26653380 -1.2547774
## t21 1.36309613 -0.8358357
## t22 1.24269972 -0.7635959
ee<-setNames(rep("blue",nrow(tree$edge)),
tree$edge[,2])
nn<-setNames(rep("grey",Ntip(tree)),1:Ntip(tree))
project.phylomorphospace(tree,X,xlab="x",ylab="y",
node.size=c(0,1.4),direction="both",nsteps=400,ftype="off",
control=list(col.edge=ee,col.node=nn),lwd=2)
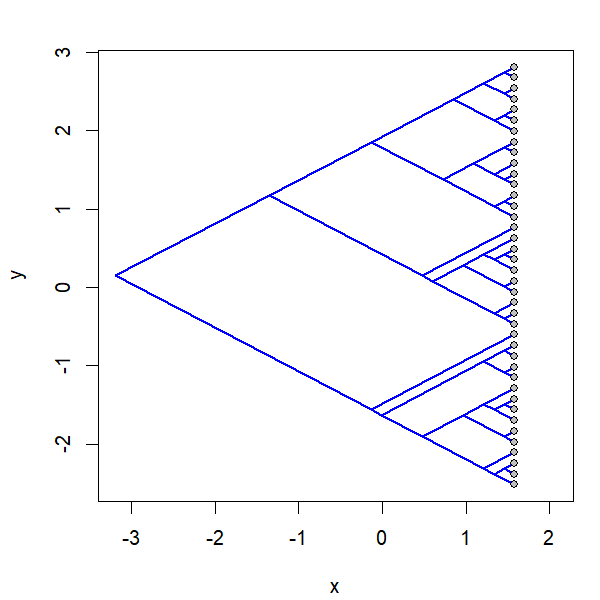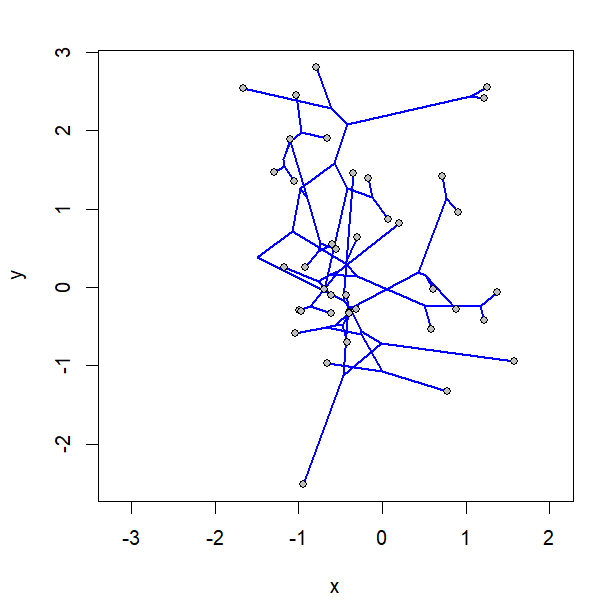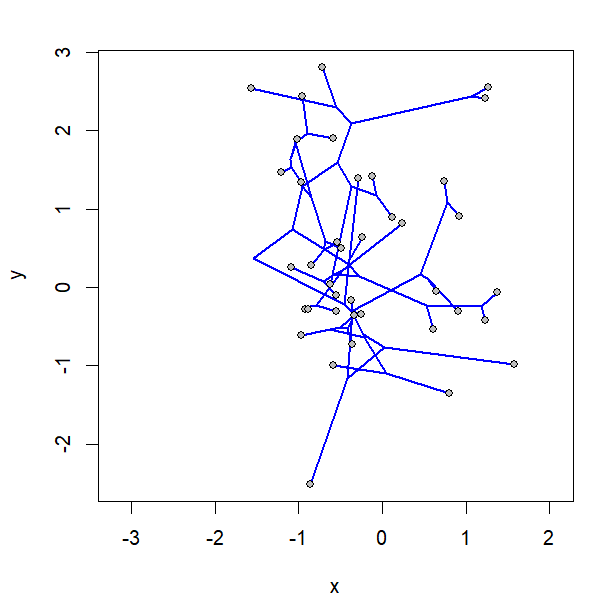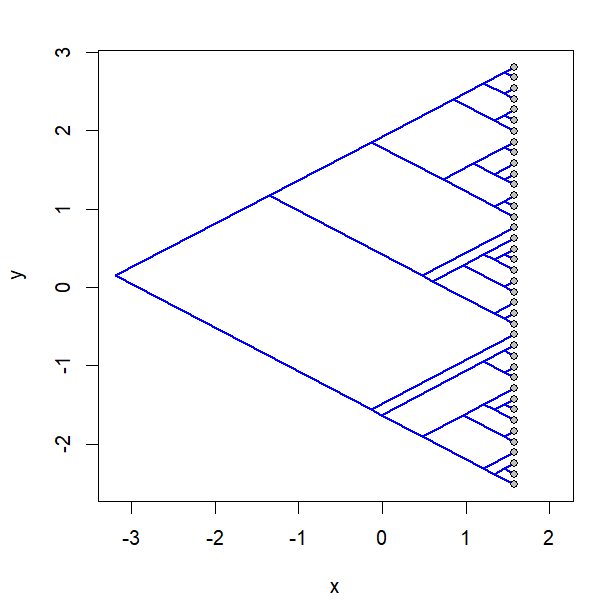
I simulated the data for x & y as follows:
tree<-pbtree(n=40)
vcv<-matrix(c(1,0,0,1),2,2)
X<-sim.corrs(tree,vcv)
and, as I've shown before, the .gif above was generated using ImageMagick which is super-handy for embedding such things into presentations & so on:
png(file="ppm-%04d.png",width=600,height=600,res=120)
par(mar=c(4.1,4.1,2.1,1.1))
project.phylomorphospace(tree,X,xlab="x",ylab="y",
node.size=c(0,1),direction="both",nsteps=100,ftype="off",
control=list(col.edge=ee,col.node=nn),lwd=2)
dev.off()
system("ImageMagick convert -delay 10 -loop 0 *.png 15Mar18-post.gif")
file.remove(list.files(pattern=".png"))