A user reports that his sample of ultrametric trees do not converge at the same end-point in a multi-tree lineage through time plot (see figure) and is perplexed why that would be.
My suspicion is that his sample is from the posterior distribution of a relaxed clock Bayesian phylogenetic analysis in BEAST. In this case (because we are sampling from the posterior distribution for the age of the root node in the tree) all the trees will have slightly different root ages and thus slightly different total lengths.
What to do with this really depends on the goal of our multi-LTT plot. If we want to represent this uncertainty, as well as the uncertainty of relative branching times, we should really make our multi-LTT plot backwards in time from a constant present-day. This is not presently implemented in phytools ltt, but is the default option (and in fact the only option that works) in ape's mltt.plot. Alternatively, and more commonly (I think), we may wish our LTT plot to primarily represent the variation in relative branching times represented in our posterior sample. To do this, then, we should first rescale all our trees to have the same length. We could rescale our trees to have an arbitrary total length (say 1 or 100), or we might want to rescale our trees to all have the same length that is the average total length of our tree. The following code does the latter:
> require(geiger); require(phytools)
> trees<-pbtree(n=100,nsim=100) # trees with different lengths
> tl<-sapply(trees,function(x) max(nodeHeights(x)))
> new.trees<-lapply(trees,rescaleTree,totalDepth=mean(tl))
> "multiPhylo"->class(new.trees)
> X<-ltt(new.trees)
And the following is a copy of the figure above with this "fix" applied:
Monday, April 30, 2012
New Bayesian method for intraspecific variation in comparative biology
My paper with Graham Reynolds is now available "Early View" from Evolution. This paper describes a new Bayesian method to account for intraspecific variation in comparative biology and is implemented in the phytools function fitBayes. Please check it out (here)!
One relatively minor frustration is that the copy editors have substituted ·, the symbol for the dot-product (an element-wise product vector sum); for ◦, the symbol for a Hadamard (element-wise matrix product). This was correct on the final version of the manuscript but I stupidly missed it in the proofs (to be fair, there were quite a few copy errors in that version). This probably does not merit a correction because I define the use of · in the text, and then it is consistent throughout. Still, slightly annoying. Oh well!
One relatively minor frustration is that the copy editors have substituted ·, the symbol for the dot-product (an element-wise product vector sum); for ◦, the symbol for a Hadamard (element-wise matrix product). This was correct on the final version of the manuscript but I stupidly missed it in the proofs (to be fair, there were quite a few copy errors in that version). This probably does not merit a correction because I define the use of · in the text, and then it is consistent throughout. Still, slightly annoying. Oh well!
Saturday, April 28, 2012
Using nlme::gls for phylogenetic regression with non-contemporaneous tips
I was just reminded tonight of the trick for performing phylogenetic regression using gls in the nlme package when the tips of the tree are non-contemporaneous. Note that there are lots of good reasons why if the tips of your tree are not contemporaneous (i.e., your tree is not ultrametric) they should be rendered so before analysis. However, there are a few different reasons (for instance, extinct species or an a priori hypothesis about how the rate of evolution of the residual error might vary among the branches of the tree) why one might wish to analyze a non-ultrametric tree.
In the function gls the correlational structure of the residual error is captured by the argument correlation. If the residual variance for different y|x differ, this must be represented in the argument weights. For a phylogenetic regression using a non-ultrametric tree this looks as follows:
> # set seed (for repeatability)
> set.seed(1)
> # simulate a tree
> tree<-rtree(n=100) # this tree is not ultrametric
> # simulate data
> x<-fastBM(tree)
> y<-0.75*x+fastBM(tree,0.2)
> # compute weights
> w<-diag(vcv.phylo(tree))
> # fit model
> fit<-gls(y~x,data=data.frame(x,y), correlation=corBrownian(1,tree),weights=varFixed(~w))
> fit
Generalized least squares fit by REML
...
Coefficients:
(Intercept) x
-1.3025017 0.7275838
...
For comparison, let's compute the contrasts regression in the typical way. This should have the same slope as our GLS regression.
> pic.x<-pic(x,tree)
> pic.y<-pic(y,tree)
> fit<-lm(pic.y~pic.x-1) # -1 is for no intercept
> fit
Call:
lm(formula = pic.y ~ pic.x - 1)
Coefficients:
pic.x
0.7276
Note that I also described this on the R-SIG-phylo email list last year (here).
In the function gls the correlational structure of the residual error is captured by the argument correlation. If the residual variance for different y|x differ, this must be represented in the argument weights. For a phylogenetic regression using a non-ultrametric tree this looks as follows:
> # set seed (for repeatability)
> set.seed(1)
> # simulate a tree
> tree<-rtree(n=100) # this tree is not ultrametric
> # simulate data
> x<-fastBM(tree)
> y<-0.75*x+fastBM(tree,0.2)
> # compute weights
> w<-diag(vcv.phylo(tree))
> # fit model
> fit<-gls(y~x,data=data.frame(x,y), correlation=corBrownian(1,tree),weights=varFixed(~w))
> fit
Generalized least squares fit by REML
...
Coefficients:
(Intercept) x
-1.3025017 0.7275838
...
For comparison, let's compute the contrasts regression in the typical way. This should have the same slope as our GLS regression.
> pic.x<-pic(x,tree)
> pic.y<-pic(y,tree)
> fit<-lm(pic.y~pic.x-1) # -1 is for no intercept
> fit
Call:
lm(formula = pic.y ~ pic.x - 1)
Coefficients:
pic.x
0.7276
Note that I also described this on the R-SIG-phylo email list last year (here).
Friday, April 27, 2012
Fixes to fitBayes and new version of phytools
Working on a different problem yesterday I discovered a relatively minor bug in the internally called MCMC functions for fitBayes, the function that implements my new Bayesian method for comparative model-fitting with intraspecific variation with Graham Reynolds (described in our 'in press' article, here). The problem was basically that when MCMC rejected a proposed new set of parameter values, it nonetheless stores the updated likelihood to memory, and returns it at the end of function execution. This is not as serious as it seems because the likelihood was not used in calculations for the next generation of the MCMC, and (especially for a long sample interval, such as the default 100 generations) will usually be numerically trivially different from the correct likelihood (although this doesn't affect our posterior sample, it could affect tests for convergence).
I have posted the new versions of these functions to my phytools page; but I have also built a new nonstatic version of phytools, to which I have also added the new version of ltt described here. You can download the source code for this new version (phytools 0.1-73) here.
I have posted the new versions of these functions to my phytools page; but I have also built a new nonstatic version of phytools, to which I have also added the new version of ltt described here. You can download the source code for this new version (phytools 0.1-73) here.
Wednesday, April 25, 2012
Simultaneously plot LTT from multiple trees in a "multiPhylo" object
Frank Burbrink asked:
is there an existing function to ltt.plot a Pp distriubtion of trees?
This already exists in the ape package (for ultrametric trees only) in the function mltt.plot. I have also added it to the phytools function ltt.
This was pretty straightforward to do, I just added code in the following general form to deal with the input of multiple trees in the form of an object of class "multiPhylo":
if(class(tree)=="multiPhylo"){
X<-lapply(tree,ltt,plot=FALSE,drop.extinct=drop.extinct, log.lineages=log.lineages,gamma=gamma)
max.lineages<-max(sapply(X,function(y) max(y$ltt)))
...
if(plot==TRUE){
...
}
return(X)
} else {
...
I have posted the code online here. I will also include it in the next version of phytools.
Just to show how it works, let's test it out:
> # load phytools & the function source
> require(phytools); source("ltt.R")
> # simulate trees
> trees<-pbtree(n=100,scale=10,nsim=100)
> # plot ltts
> Z<-ltt(trees,log=F)
Pretty cool. The function also returns the branching times and number of lineages, as well as values for Pybus & Harvey's (2000) γ-statistic & P value. So, for instance, if we want to plot a histogram showing the distribution of γ among trees we can just do:
> gamma<-sapply(Z,function(y) y$gamma)
> hist(gamma)
Or if we would like to calculate the number of significant values of γ in our set:
> nsig<-sum(sapply(Z,function(y) y$p<0.05))
> nsig
[1] 3
Cool.
is there an existing function to ltt.plot a Pp distriubtion of trees?
This already exists in the ape package (for ultrametric trees only) in the function mltt.plot. I have also added it to the phytools function ltt.
This was pretty straightforward to do, I just added code in the following general form to deal with the input of multiple trees in the form of an object of class "multiPhylo":
if(class(tree)=="multiPhylo"){
X<-lapply(tree,ltt,plot=FALSE,drop.extinct=drop.extinct, log.lineages=log.lineages,gamma=gamma)
max.lineages<-max(sapply(X,function(y) max(y$ltt)))
...
if(plot==TRUE){
...
}
return(X)
} else {
...
I have posted the code online here. I will also include it in the next version of phytools.
Just to show how it works, let's test it out:
> # load phytools & the function source
> require(phytools); source("ltt.R")
> # simulate trees
> trees<-pbtree(n=100,scale=10,nsim=100)
> # plot ltts
> Z<-ltt(trees,log=F)
Pretty cool. The function also returns the branching times and number of lineages, as well as values for Pybus & Harvey's (2000) γ-statistic & P value. So, for instance, if we want to plot a histogram showing the distribution of γ among trees we can just do:
> gamma<-sapply(Z,function(y) y$gamma)
> hist(gamma)
Or if we would like to calculate the number of significant values of γ in our set:
> nsig<-sum(sapply(Z,function(y) y$p<0.05))
> nsig
[1] 3
Cool.
Tuesday, April 24, 2012
Adding the expectation to a LTT plot
A user writes:
how do I add the a line with the expected number of lineages to a semi-log lineage-through-time plot?
The answer is simple enough, just use the base function lines, for example:
> require(phytools)
> tree<-pbtree(n=100)
> z<-ltt(tree)
> lines(c(0,max(nodeHeights(tree))),c(log(2), log(length(tree$tip))),lty=2)
That's it.
how do I add the a line with the expected number of lineages to a semi-log lineage-through-time plot?
The answer is simple enough, just use the base function lines, for example:
> require(phytools)
> tree<-pbtree(n=100)
> z<-ltt(tree)
> lines(c(0,max(nodeHeights(tree))),c(log(2), log(length(tree$tip))),lty=2)
That's it.
Monday, April 23, 2012
Error in the estimation of species means and model selection in comparative biology
This will also be part of the focus of my upcoming symposium talk at Evolution this summer, but it is becoming my impression that there is a distinct lack of appreciation for how error in the estimation of species means can affect model selection in comparative biology. Error in the estimation of species means is undoubtedly pervasive. For example, authors commonly measure 1-10 specimens per species, meaning that (if intraspecific variability is modest or high), sampling error can be substantial.
Just to give two quick examples, let's consider the consequences of (ignored) error in the estimation of species means for a single-optimum OU model (Ornstein-Uhlenbeck), and an EB (Early-burst) model.
First, let's simulate tree & data under Brownian evolution:
> require(phytools); require(geiger)
> set.seed(10) # for repeatability, but try multiple seeds
> tree<-pbtree(n=200,scale=1)
> x<-fastBM(tree)
Now, let's add some sampling error using the function sampleFrom in phytools:
> xe<-sampleFrom(x,0.2,rep(1,200))
We can easily see that x and xe are quite closely correlated - about as correlated as we might expect our true & estimated species means to be in any empirical study:
> plot(x,xe)
Next, let's try to fit BM & OU models to our data:
> fitBM<-fitContinuous(tree,xe,model="BM")
Fitting BM model:
> fitBM
$Trait1
$Trait1$lnl
[1] -348.0691
$Trait1$beta
[1] 9.366567
$Trait1$aic
[1] 700.1382
...
> fitOU<-fitContinuous(tree,xe,model="OU")
Fitting OU model:
> fitOU
$Trait1
$Trait1$lnl
[1] -279.5255
$Trait1$beta
[1] 126.3536
$Trait1$alpha
[1] 63.67358
...
$Trait1$aic
[1] 565.051
...
(It can easily be verified that this is not true of the original data vector x, for which BM is the better model:
> fitContinuous(tree,x,model="BM")$Trait1$aic < fitContinuous(tree,x,model="OU")$Trait1$aic
Fitting BM model:
Fitting OU model:
[1] TRUE ).
We can see already that for a good amount of sampling error in the estimation of species means, our fitted model flips - from BM (the generating model) to OU. We can also test this for EB. Here, I will adjust the bounds on alpha, to allow either a decrease or an increase in the evolutionary rate through time.
> fitEB<-fitContinuous(tree,xe,model="EB", bounds=list(alpha=c(-100,100)))
Fitting EB model:
> fitEB
$Trait1
$Trait1$lnl
[1] -282.1674
$Trait1$beta
[1] 1e-08
$Trait1$a
[1] 21.63922
$Trait1$aic
[1] 570.3348
...
Here again we see that EB fits much better than the generating model, BM. Of course, it is possible to address this bias by following the method of Ives et al. (2007), and this is even implemented in fitContinuous. Let's try again for the EB example: > fitBMe<-fitContinuous(tree,xe,model="BM", meserr=rep(sqrt(0.2),200))
Fitting BM model:
> fitBMe
$Trait1
$Trait1$lnl
[1] -218.4676
$Trait1$beta
[1] 0.908003
$Trait1$aic
[1] 440.9353
$Trait1$aicc
[1] 440.9965
$Trait1$k
[1] 2
> fitEBe<-fitContinuous(tree,xe,model="EB", bounds=list(alpha=c(-100,100)),meserr=rep(0.2,200))
Fitting EB model:
> fitEBe
$Trait1
$Trait1$lnl
[1] -218.4567
$Trait1$beta
[1] 0.8149614
$Trait1$a
[1] 0.1468192
$Trait1$aic
[1] 442.9134
$Trait1$aicc
[1] 443.0365
$Trait1$k
[1] 3
Cool. This is what we might expect. Finally, it is interesting to consider what might happen if phenotypic data actually arose under EB, but then sampling error in the estimation of species means is (again) ignored.
> x<-fastBM(linearchangeTree(tree,slope=-0.5))
> xe<-sampleFrom(x,0.2,rep(1,200))
> fitContinuous(tree,x,model="EB")
Fitting EB model:
$Trait1
$Trait1$lnl
[1] -69.14438
$Trait1$beta
[1] 0.9732424
$Trait1$a
[1] -0.6107059
$Trait1$aic
[1] 144.2888
$Trait1$aicc
[1] 144.4118
$Trait1$k
[1] 3
> fitContinuous(tree,xe,model="EB")
Fitting EB model:
$Trait1
$Trait1$lnl
[1] -270.8313
$Trait1$beta
[1] 4.326543 $Trait1$a
[1] 0
$Trait1$aic
[1] 547.6626
$Trait1$aicc
[1] 547.7857
$Trait1$k
[1] 3
In this example, a very strong EB pattern vanishes when there is sampling error.
Of course, the reader should keep in mind that all of the above results depend critically on the size of sampling error relative to the variance among species. If this is very small, it can be much more safely ignored. For example:
> xe<-sampleFrom(x,0.001,rep(1,200))
> fitContinuous(tree,xe,model="EB")
Fitting EB model:
$Trait1
$Trait1$lnl
[1] -75.14334
$Trait1$beta
[1] 0.8179839
$Trait1$a
[1] -0.3384102
$Trait1$aic
[1] 156.2867
$Trait1$aicc
[1] 156.4098
$Trait1$k
[1] 3
As I mentioned above, a more detailed treatment of this and other issues is underway for my SSB symposium talk at Evolution 2012 in Ottawa.
Just to give two quick examples, let's consider the consequences of (ignored) error in the estimation of species means for a single-optimum OU model (Ornstein-Uhlenbeck), and an EB (Early-burst) model.
First, let's simulate tree & data under Brownian evolution:
> require(phytools); require(geiger)
> set.seed(10) # for repeatability, but try multiple seeds
> tree<-pbtree(n=200,scale=1)
> x<-fastBM(tree)
Now, let's add some sampling error using the function sampleFrom in phytools:
> xe<-sampleFrom(x,0.2,rep(1,200))
We can easily see that x and xe are quite closely correlated - about as correlated as we might expect our true & estimated species means to be in any empirical study:
> plot(x,xe)
Next, let's try to fit BM & OU models to our data:
> fitBM<-fitContinuous(tree,xe,model="BM")
Fitting BM model:
> fitBM
$Trait1
$Trait1$lnl
[1] -348.0691
$Trait1$beta
[1] 9.366567
$Trait1$aic
[1] 700.1382
...
> fitOU<-fitContinuous(tree,xe,model="OU")
Fitting OU model:
> fitOU
$Trait1
$Trait1$lnl
[1] -279.5255
$Trait1$beta
[1] 126.3536
$Trait1$alpha
[1] 63.67358
...
$Trait1$aic
[1] 565.051
...
(It can easily be verified that this is not true of the original data vector x, for which BM is the better model:
> fitContinuous(tree,x,model="BM")$Trait1$aic < fitContinuous(tree,x,model="OU")$Trait1$aic
Fitting BM model:
Fitting OU model:
[1] TRUE ).
We can see already that for a good amount of sampling error in the estimation of species means, our fitted model flips - from BM (the generating model) to OU. We can also test this for EB. Here, I will adjust the bounds on alpha, to allow either a decrease or an increase in the evolutionary rate through time.
> fitEB<-fitContinuous(tree,xe,model="EB", bounds=list(alpha=c(-100,100)))
Fitting EB model:
> fitEB
$Trait1
$Trait1$lnl
[1] -282.1674
$Trait1$beta
[1] 1e-08
$Trait1$a
[1] 21.63922
$Trait1$aic
[1] 570.3348
...
Here again we see that EB fits much better than the generating model, BM. Of course, it is possible to address this bias by following the method of Ives et al. (2007), and this is even implemented in fitContinuous. Let's try again for the EB example: > fitBMe<-fitContinuous(tree,xe,model="BM", meserr=rep(sqrt(0.2),200))
Fitting BM model:
> fitBMe
$Trait1
$Trait1$lnl
[1] -218.4676
$Trait1$beta
[1] 0.908003
$Trait1$aic
[1] 440.9353
$Trait1$aicc
[1] 440.9965
$Trait1$k
[1] 2
> fitEBe<-fitContinuous(tree,xe,model="EB", bounds=list(alpha=c(-100,100)),meserr=rep(0.2,200))
Fitting EB model:
> fitEBe
$Trait1
$Trait1$lnl
[1] -218.4567
$Trait1$beta
[1] 0.8149614
$Trait1$a
[1] 0.1468192
$Trait1$aic
[1] 442.9134
$Trait1$aicc
[1] 443.0365
$Trait1$k
[1] 3
Cool. This is what we might expect. Finally, it is interesting to consider what might happen if phenotypic data actually arose under EB, but then sampling error in the estimation of species means is (again) ignored.
> x<-fastBM(linearchangeTree(tree,slope=-0.5))
> xe<-sampleFrom(x,0.2,rep(1,200))
> fitContinuous(tree,x,model="EB")
Fitting EB model:
$Trait1
$Trait1$lnl
[1] -69.14438
$Trait1$beta
[1] 0.9732424
$Trait1$a
[1] -0.6107059
$Trait1$aic
[1] 144.2888
$Trait1$aicc
[1] 144.4118
$Trait1$k
[1] 3
> fitContinuous(tree,xe,model="EB")
Fitting EB model:
$Trait1
$Trait1$lnl
[1] -270.8313
$Trait1$beta
[1] 4.326543 $Trait1$a
[1] 0
$Trait1$aic
[1] 547.6626
$Trait1$aicc
[1] 547.7857
$Trait1$k
[1] 3
In this example, a very strong EB pattern vanishes when there is sampling error.
Of course, the reader should keep in mind that all of the above results depend critically on the size of sampling error relative to the variance among species. If this is very small, it can be much more safely ignored. For example:
> xe<-sampleFrom(x,0.001,rep(1,200))
> fitContinuous(tree,xe,model="EB")
Fitting EB model:
$Trait1
$Trait1$lnl
[1] -75.14334
$Trait1$beta
[1] 0.8179839
$Trait1$a
[1] -0.3384102
$Trait1$aic
[1] 156.2867
$Trait1$aicc
[1] 156.4098
$Trait1$k
[1] 3
As I mentioned above, a more detailed treatment of this and other issues is underway for my SSB symposium talk at Evolution 2012 in Ottawa.
Common error in make.simmap
A user today reported encountering the following error while using the phytools function make.simmap:
> mtree<-make.simmap(tree,y)
Error in if (names(map)[length(map)] == node.states[j, 2]) accept = TRUE :
missing value where TRUE/FALSE needed
In addition: Warning message:
In sqrt(diag(solve(h))) : NaNs produced
It turns out that this is just due to the fact that the data y is a data frame, with species names in rows, rather than a vector with names(...) equal to the species names. Perhaps in the future I will modify make.simmap to rectify this problem, but for no this can be circumvented as follows:
> z<-y; names(z)<-rownames(y)
> mtree<-make.simmap(tree,z)
I figured that this was probably a sufficiently common problem for users of make.simmap to repeat on the blog.
> mtree<-make.simmap(tree,y)
Error in if (names(map)[length(map)] == node.states[j, 2]) accept = TRUE :
missing value where TRUE/FALSE needed
In addition: Warning message:
In sqrt(diag(solve(h))) : NaNs produced
It turns out that this is just due to the fact that the data y is a data frame, with species names in rows, rather than a vector with names(...) equal to the species names. Perhaps in the future I will modify make.simmap to rectify this problem, but for no this can be circumvented as follows:
> z<-y; names(z)<-rownames(y)
> mtree<-make.simmap(tree,z)
I figured that this was probably a sufficiently common problem for users of make.simmap to repeat on the blog.
Thursday, April 19, 2012
Comparative methods in R workshop
 Just a quick mention that Luke Harmon (University of Idaho) & Mike Alfaro (UCLA) are once again offering their very popular workshop on macroevolutionary analysis in the R environment. By all accounts this is a great course that should be of considerable interest to readers of this blog. To sweeten the deal, there are even a limited number of competitive scholarships available to defray costs of travel, room, & board for some applicants. Note that I have no direct involvement in this course.
Just a quick mention that Luke Harmon (University of Idaho) & Mike Alfaro (UCLA) are once again offering their very popular workshop on macroevolutionary analysis in the R environment. By all accounts this is a great course that should be of considerable interest to readers of this blog. To sweeten the deal, there are even a limited number of competitive scholarships available to defray costs of travel, room, & board for some applicants. Note that I have no direct involvement in this course.For more information contact Luke or Mike. I have also re-posted the full course announcement below:
**************************************************************
Macroevolution in R Short Course
We are pleased to announce an intensive short course on using R to perform comparative methods to be held in Santa Barbara on June 11th to June 15th. This course is funded by the National Science Foundation, and a number of stipends to cover or defray travel, room, and board are available to qualified students and post-docs. Topics covered will include an introduction to the R programming language, tree manipulation, independent contrasts and phylogenetic generalized least squares, ancestral state reconstruction, models of character evolution, diversification analyses, and community phylogenetic analysis. If you are interested please submit your CV along with a short (maximum 1 page) description of your research interests, background, and reasons for taking the course. Admission is competitive, and the best applications come from students with data sets to analyze. International applicants are welcome.
To apply visit this URL: tinyurl.com/macro-in-R
Application deadline: April 30th.
Individuals from cultural, racial, linguistic, geographic and socioeconomic backgrounds that are currently underrepresented in science are especially encouraged to apply.
Please contact the co-organizers, Michael Alfaro (michaelalfaro@ucla.edu) and Luke Harmon (lukeh@uidaho.edu) with any questions.
**************************************************************
Wednesday, April 11, 2012
phytools article published
 I just discovered that the phytools article has been officially published in Methods in Ecology & Evolution. The article, like all program notes published in MEE, is available "open access" (link to article here). Check it out.
I just discovered that the phytools article has been officially published in Methods in Ecology & Evolution. The article, like all program notes published in MEE, is available "open access" (link to article here). Check it out.The article is already "out of date," in the sense that its list of phytools functionality leaves out many new features in the package, nonetheless some users might find the worked examples (which includes vignette style code from an interactive R session) helpful in guiding their use of some of phytools' functions.
Monday, April 9, 2012
Facebook page for Puerto Rico course
 I just created a Facebook page for the tropical biology course in Puerto Rico that I will be teaching, starting in January of 2013, in collaboration with Alberto Puente-Rolón. The course is a three week field-based course that will visit all of the major ecosystems of the Caribbean. The URL for the Facebook page as follows: http://www.facebook.com/TropicalBiologyInPuertoRico. So far the page includes only an abbreviated course description and some additional photos, but I will add details, photos, and links as they develop. Please check it out, "like" us, or share with your friends.
I just created a Facebook page for the tropical biology course in Puerto Rico that I will be teaching, starting in January of 2013, in collaboration with Alberto Puente-Rolón. The course is a three week field-based course that will visit all of the major ecosystems of the Caribbean. The URL for the Facebook page as follows: http://www.facebook.com/TropicalBiologyInPuertoRico. So far the page includes only an abbreviated course description and some additional photos, but I will add details, photos, and links as they develop. Please check it out, "like" us, or share with your friends.The photo above, by the way, was taken from the south coast of the island by Brian Langerhans on a field trip by he & I down there in 2005.
Saturday, April 7, 2012
New package for population genetic simulation & numerical analysis
I have just posted a new package, 'popgen', that does some relatively simple numerical analyses of basic population genetic models, as well as genetic drift and founder effect simulations. These are mostly a small number of functions that I have developed for teaching the population genetics section of the undergraduate evolutionary biology class I'm giving this semester (as well as one function that I developed for a paper I have submitted in collaboration with Manuel Leal at Duke). Consequently, the functions generally generate animations (e.g., here). I have usually incorporated a few different options for plotting in each case. For instance, the two figures below illustrate the end result of the same simulation of genetic drift in 10 populations for the allele frequency, p, and the mean heterozygosity (in the latter case, the red curved line shows the expected decline in heterozygosity through time). It is not currently possible to generate both animations at once; rather to replicate the same simulation and plot different aspects (gene frequency, genotype frequencies, heterozygosity, etc.) is necessary to control the seed and re-run. For instance, in the cases below I just did the following:
> require(popgen)
> set.seed(1); genetic.drift() # using all defaults
> set.seed(1); genetic.drift(show="heterozygosity")
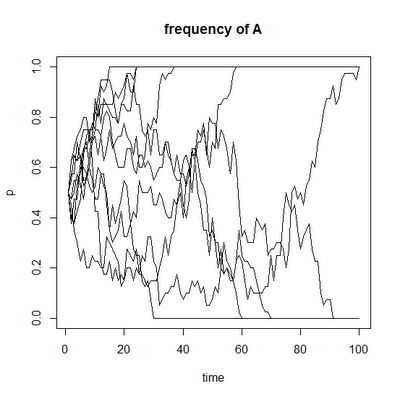

I mostly built this package to make it easier to save, document & load the code I have developed for future instances in which I might teach this course - but now that I have done so, I thought that I might as well share it with the world!
A direct link to the package is here after which you can install as follows:
> install.packages("popgen_0.1.tar.gz",type="source",repos=NULL)
The package is also linked of my programs page where there is also a link to a PDF manual for the package.
> require(popgen)
> set.seed(1); genetic.drift() # using all defaults
> set.seed(1); genetic.drift(show="heterozygosity")


I mostly built this package to make it easier to save, document & load the code I have developed for future instances in which I might teach this course - but now that I have done so, I thought that I might as well share it with the world!
A direct link to the package is here after which you can install as follows:
> install.packages("popgen_0.1.tar.gz",type="source",repos=NULL)
The package is also linked of my programs page where there is also a link to a PDF manual for the package.
New article out "Accepted" in Evolution
Just a quick comment to note that my new paper with Graham Reynolds has just come out "Accepted" (that is, in manuscript form in advance of publication) in Evolution. The paper describes a new Bayesian method to incorporate intraspecific data in phylogenetic comparative analyses. A link to the article can be found here. Check it out! (And if you don't have access, please let me know and I would be happy to send you a PDF.)
Tuesday, April 3, 2012
Field biology course in Puerto Rico
 This is neither phylogenetics nor 'phytools' related, at least not directly, but I just wanted to take this opportunity to announce a three week, field-based course in tropical biology - focusing on ecology, evolution, and conservation biology - to be offered for the first time in January 2013 during the UMass Boston winter session (and hopefully annually thereafter). A more detailed description of the course can be found on the UMass Boston international programs page, here. The course will be co-instructed by myself and Alberto Puente-Rolón, professor at the Universidad Interamericana de Puerto Rico, Arecibo campus, with guest instruction for at least part of the course by Graham Reynolds. Alberto, it should be noted, took both of the photos on the international programs page, as well as the photo (of Anolis evermanni) shown above. The price for the course (determined by the university and listed on the program webpage) includes all travel to, from, and within the island, all food, and all accommodation (along with tuition and fees, of course). We also hope to reduce this depending on enrollment numbers and our final budget for the course.
This is neither phylogenetics nor 'phytools' related, at least not directly, but I just wanted to take this opportunity to announce a three week, field-based course in tropical biology - focusing on ecology, evolution, and conservation biology - to be offered for the first time in January 2013 during the UMass Boston winter session (and hopefully annually thereafter). A more detailed description of the course can be found on the UMass Boston international programs page, here. The course will be co-instructed by myself and Alberto Puente-Rolón, professor at the Universidad Interamericana de Puerto Rico, Arecibo campus, with guest instruction for at least part of the course by Graham Reynolds. Alberto, it should be noted, took both of the photos on the international programs page, as well as the photo (of Anolis evermanni) shown above. The price for the course (determined by the university and listed on the program webpage) includes all travel to, from, and within the island, all food, and all accommodation (along with tuition and fees, of course). We also hope to reduce this depending on enrollment numbers and our final budget for the course.If you have any questions about the course or would be willing to advertise it at your home institution, please contact me. Keep in mind that due to variation in winter break times at different universities the period of this course may conflict with the start of spring semester for many students.
Fix for problem with anc.Bayes
A user reports the following problem (here):
I am using your anc.Bayes function in phytools with a nexus tree and continuous data. When I run the line "anc.Bayes(tree,data,ngen=10000,control=list())" I get an error message:
"Error in if (post.odds > runif(n = 1)) { : missing value where TRUE/FALSE needed"
I still have not identified the specific cause of this error, but the problem seems to be fixed by converting the input data (originally stored as a data frame) into a vector. Of course, as always, one has to be careful to preserve the species names which were row names in the data frame, and must be names in the vector.
I.e.,
# x is a data frame
x<-as.matrix(x)[,1] # now x is a named vector
result<-anc.Bayes(tree,x) # for instance
I hope this helps.
I am using your anc.Bayes function in phytools with a nexus tree and continuous data. When I run the line "anc.Bayes(tree,data,ngen=10000,control=list())" I get an error message:
"Error in if (post.odds > runif(n = 1)) { : missing value where TRUE/FALSE needed"
I still have not identified the specific cause of this error, but the problem seems to be fixed by converting the input data (originally stored as a data frame) into a vector. Of course, as always, one has to be careful to preserve the species names which were row names in the data frame, and must be names in the vector.
I.e.,
# x is a data frame
x<-as.matrix(x)[,1] # now x is a named vector
result<-anc.Bayes(tree,x) # for instance
I hope this helps.
Subscribe to:
Posts (Atom)






