The post title pretty much says it all.
The only challenging thing here was dealing with time-stops (we don't know how many tips we'll end up with, so we can't specify tip labels a priori) and taxa-stop with extinction (we define this based on the number of extant lineages - if we choose to also return extinct tips, what do we call them?).
Code for the new version of the function is here, and it is in a new phytools build (phytools 0.3-49) that can be installed from source.
Here's a demo:
> library(phytools)
> packageVersion("phytools")
[1] ‘0.3.49’
> tree<-pbtree(n=26,tip.label=LETTERS[26:1])
> plotTree(tree)
> packageVersion("phytools")
[1] ‘0.3.49’
> tree<-pbtree(n=26,tip.label=LETTERS[26:1])
> plotTree(tree)
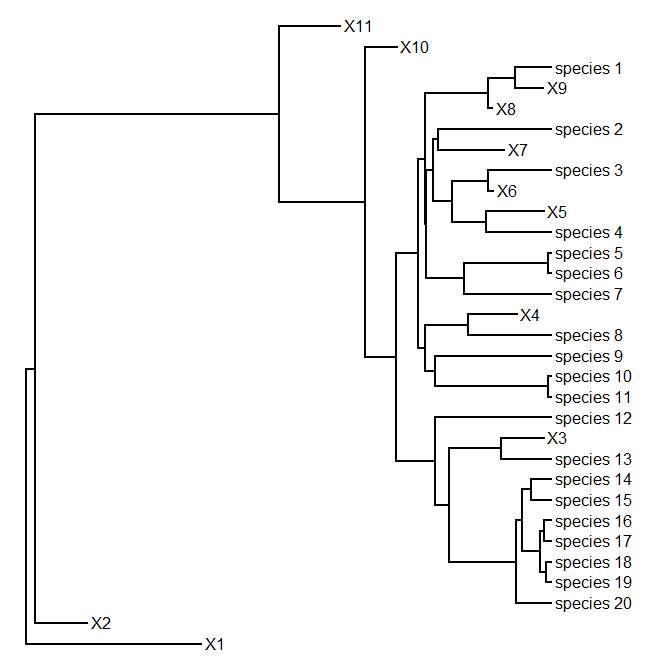
> tree<-pbtree(b=1,d=0.3,n=20,tip.label=paste("species_", 20:1,sep=""))
Warning: only using labels in tip.label for extant tips.
extinct tips will be labeled X1, X2, etc.
> plotTree(tree)
Warning: only using labels in tip.label for extant tips.
extinct tips will be labeled X1, X2, etc.
> plotTree(tree)
> ## this will probably go completely extinct
> tree<-pbtree(b=1,d=0.9,n=10,tip.label=letters[1:10])
Warning: only using labels in tip.label for extant tips.
extinct tips will be labeled X1, X2, etc.
> plotTree(tree)
> tree<-pbtree(b=1,d=0.9,n=10,tip.label=letters[1:10])
Warning: only using labels in tip.label for extant tips.
extinct tips will be labeled X1, X2, etc.
> plotTree(tree)
> ## n & t
> n<-26
> b<-1
> d<-0.4
> t<-log(26/2)/(b-d)
> tree<-pbtree(n=n,t=t,b=b,d=d,tip.label=LETTERS[26:1], method="direct")
Warning: only using labels in tip.label for extant tips.
extinct tips will be labeled X1, X2, etc.
simulating with both taxa-stop (n) & time-stop (t) using
'direct' sampling. this is experimental
> plotTree(tree)
> n<-26
> b<-1
> d<-0.4
> t<-log(26/2)/(b-d)
> tree<-pbtree(n=n,t=t,b=b,d=d,tip.label=LETTERS[26:1], method="direct")
Warning: only using labels in tip.label for extant tips.
extinct tips will be labeled X1, X2, etc.
simulating with both taxa-stop (n) & time-stop (t) using
'direct' sampling. this is experimental
> plotTree(tree)



No comments:
Post a Comment
Note: due to the very large amount of spam, all comments are now automatically submitted for moderation.