I just added a new function cladelabels to the phytools package. This function is in some ways analogous to nodelabels, tiplabels, etc. in ape. It basically implements the method I gave here, while taking advantage of the trick described here.
The code for this function is here, and it is also in a new phytools version, which can be downloaded here.
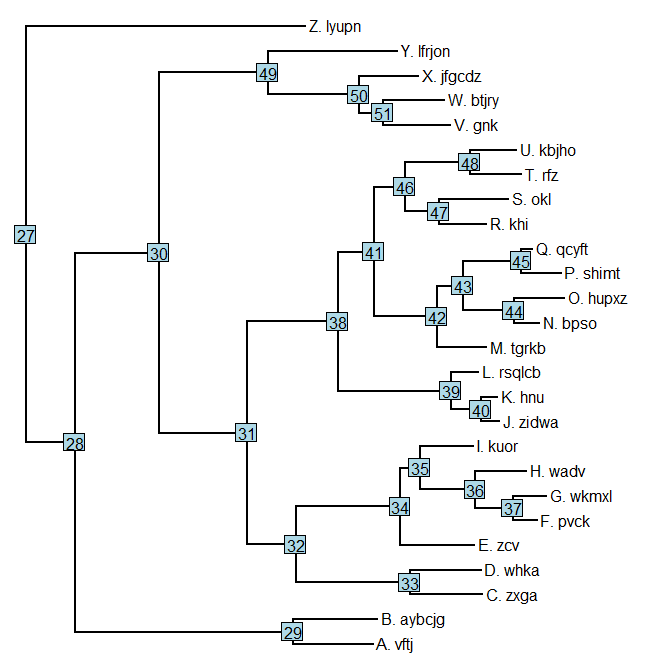
Here's a demo:
tree<-pbtree(n=26,tip.label=
paste(LETTERS,"._",sapply(round(runif(n=26,min=3,max=6)),
function(x) paste(sample(letters,x),collapse="")),sep=""),
scale=1)
tree$edge.length<-tree$edge.length+0.1*rchisq(n=
length(tree$edge.length),df=10)/10
plotTree(tree) ## also can use plot.phylo
nodelabels()
Now let's label the three clades descended from nodes 46 & 33; and then also node 28 (which is inclusive of node 33:
plotTree(tree,xlim=c(0,1.25*max(nodeHeights(tree))),
ftype="i")
cladelabels(tree,node=49,"Clade A")
cladelabels(tree,node=34,"Clade B")
cladelabels(tree,node=31,"Clade C",offset=3))
Cool - this was more or less what we were going for. We can also do this without sending cladelabels the tree, although in this case we need to provide some guidance on the space that cladelabels should leave for tip labels - otherwise it will assume a fixed width of 8 characters.
ftype="i")
## delete the tree! (we really don't need it)
rm(tree)
cladelabels(node=49,text="Clade A",offset=4.5)
cladelabels(node=34,text="Clade B",offset=6)
cladelabels(node=31,text="Clade C",offset=9)
It's also fairly obvious how this could be combined with findMRCA to label clades on the basis of the taxa in the clade, rather than their specific MRCA node number. For instance:
ftype="i")
tips<-c("L._rsqlcb","J._zidwa","K._hnu","M._tgrkb",
"N._bpso","O._hupxz","P._shimt","Q._qcyft",
"R._khi","S._okl","T._rfz","U._kbjho")
cladelabels(tree,node=findMRCA(tree,tips),text="clade D",
offset=2)
Finally, it's possible to send the function multiple node numbers & clade labels in one function call - although at present this does not permit us to use different offset values. So:
cladelabels(tree,node=34,"Clade B")
## is the same as:
cladelabels(tree,node=c(49,34),text=c("Clade A","Clade B"))
One caveat important to mention is that at present this works only for rightward facing phylograms (or cladograms, for tree=NULL). This is not theoretically difficult to extend to other plot types, it just requires more work.



Hey Liam,
ReplyDeleteThis seems like a really nice and easy way of doing exactly what I need to do. Only problem is that R cannot find the cladelabel function?
I installed the latest version of the phytools package but it doesn't seem to work...
Any idea?
Cheers,
Luca
This is not yet on CRAN. You need to install from source after downloading from here. Let me know if you are not able to install from source & can post a Windows binary version. (At this time I cannot post a Mac OS X binary version.) -- Liam
DeleteHi Liam,
DeleteSorry, my bad. Just downloaded it and works perfectly!
Thanks very much.
Would be great if it could be modified for type="fan" too ;-)
Thanks again,
Luca