I just submitted a new version (Rphylip 0.1-23) of the Rphylip package ('an R interface for PHYLIP') to CRAN. This version fixes a couple of bugs in the first CRAN version - including taxon name length limits in Rconsense and Rtreedist (which are present in the corresponding PHYLIP programs CONSENSE and TREEDIST, but can easily be circumvented when calling PHYLIP from within R). It also introduces the new function Rseqboot, which is an R interface two SEQBOOT. SEQBOOT implements a range of non-parametric bootstrapping, jacknife, and data permutation methods. Because it can take a variety of different character types as input, writing the interface was a bit of a pain in the butt - but it is finished to my satisfaction today.
Here's a demo of bootstrapping, distance matrix calculation from all bootstrap samples, and then consensus phylogeny estimation using the Rphylip package:
Loading required package: Rphylip
> packageVersion("Rphylip")
[1] ‘0.1.23’
> ## load primate dataset
> data(primates)
> ## basic bootstrapping
> X<-Rseqboot(primates,quiet=TRUE)
> ## compute distance matrices
> D<-lapply(X,Rdnadist,quiet=TRUE)
> ## compute all NJ trees
> ## note Rneighbor(...,quiet=TRUE) is
> ## not as quiet as it should be!
> trees<-lapply(D,Rneighbor,quiet=TRUE)
> ## reroot all trees using midpoint rooting
> require(phangorn)
Loading required package: phangorn
> trees<-lapply(trees,midpoint)
> class(trees)<-"multiPhylo"
> ## compute consensus tree
> tree<-Rconsense(trees,quiet=TRUE,rooted=TRUE)
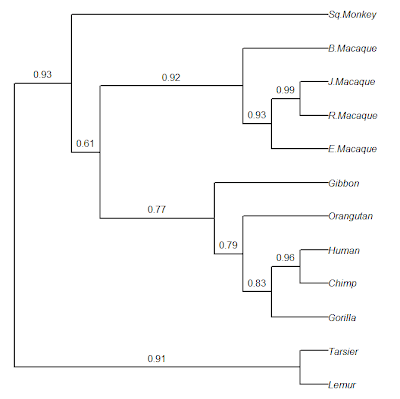
> ## now plot the result with the bootstrap %s
> plot(tree,edge.width=2,no.margin=TRUE)
> ## find edges
> e<-sapply(2:tree$Nnode+length(tree$tip.label),
function(x,y) which(y==x),y=tree$edge[,2])
> ## add bootstrap
> edgelabels(tree$node.label[2:tree$Nnode],e,pos=3,
frame="none",bg="transparent")
Cool. Note that RETREE in PHYLIP does midpoint rooting, but this program cannot yet be called with Rphylip. The same general pipeline could be used with ML, MP, or other phylogeny inference methods in PHYLIP (although this would be slower, of course).
This version of Rphylip is already available from GitHub, but hopefully will also be accepted on CRAN soon.
I am rewriting notes on a graduate plylogenetics course, using the PHYLIP suite. The predecessor had the original command line phylip demonstrated and the comparison with the elegance of the code in R, and how much faster and automated it becomes, is so obvious. Your functions are such a vast imrovement on the process, I am more than happy to be able to demonstrate this to the students. Really kind of you to implement these features, Liam! Thank you!
ReplyDeleteI tried the bootstrap approach, above, using a parsimony program (dollop). However, since parsimony programs will often output several trees, the corresponding 'trees' object (to that in your example) from my bootstrap is a list of a list of phylo instances, which Rconsense cannot handle. Moreover, it would, in this case, be preferable to down-weight trees from any specific bootstrap replicate, bu 1/(number of trees from that bootstrap) to get the estimated bootstrap fractions correct. Do you have any suggestions on how to handle this or am I missing something obvious? Thanks!
ReplyDelete