To illustrate this, I can first simulate the tree & data:
> require(phytools); require(geiger)
> set.seed(1)
> tree<-rbdtree(b=30,d=0,Tmax=0.1)
> x<-sim.char(tree,model.matrix=list(matrix(c(-10,10,10,-10),2,2)), model="discrete")[,,1]
> mtree<-make.simmap(tree,x)
> cols<-c("blue","red"); names(cols)<-c(1,2)
> plotSimmap(mtree,fsize=0.7,cols)
Which produces the following result:

(border added for illustrative effect).
I had the hardest time trying to resolve this issue; however I finally realized that if I first rescaled the sum of the total tree length and the maximum string width of the longest taxon name (which can be found using strwidth(), once a plotting window has been opened) to 1.0, then the problem is solved.
Let's load the new version of plotSimmap() and try again:
> source("http://faculty.umb.edu/liam.revell/phytools/plotSimmap/v0.6/plotSimmap.R")
> plotSimmap(mtree,fsize=0.7,cols)
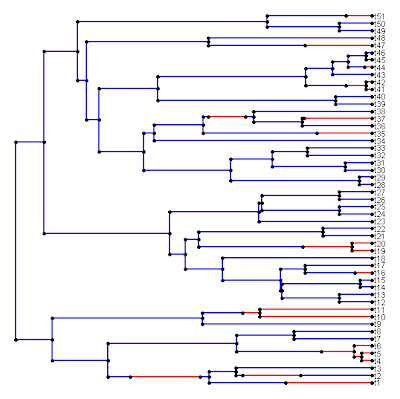
Indeed, this looks much better.
Direct link to code for this function is here. I have only tried this on one system - so I would be happy to hear reports that it works on other machines (particularly Mac OS or Linux).
No comments:
Post a Comment
Note: due to the very large amount of spam, all comments are now automatically submitted for moderation.