Recently an R-sig-phylo subscriber made the following request:
"I need to add a list of 230 species in a phylogenetic tree. Is there a logical way to add the species to the root of
the genera to which it belongs, in a systematic way, that is, to make a function recognizes the name of the genera to which the species belongs, and so it adds the species to that root?"
I posted a solution to this, but I also thought that other phytools users might face the same problem so I have now automated this in the function add.species.to.genus. This, along with the matrix comparison function skewers, is also in a new minor phytools build (phytools 0.3-73).
The function works by first peeling the genus name out of the species name. It does this by looking for either the underscore character, "_", or a space character, " ". It then proceeds to identify the clade containing con-generics in the tree by matching the genus name to the tip labels. Finally, it can either attach the new species to the root node of the most inclusive clade containing congenerics; or it can attach the new species randomly within that clade. In general, the function works best if the input tree is ultrametric. Otherwise, it may return a tree without edge lengths!
Here's a quick demo:
> library(phytools)
Loading required package: ape
Loading required package: maps
Loading required package: rgl
> packageVersion("phytools")
[1] ‘0.3.73’
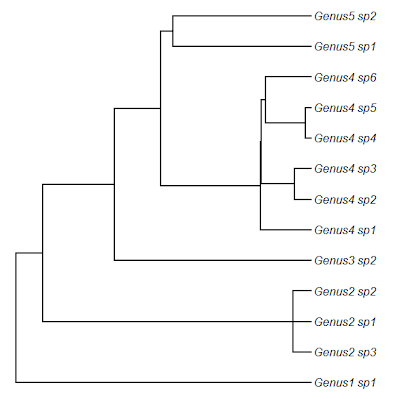
> ## here's our starting tree
> plotTree(tree,ftype="i")
> species<-"Genus2_sp3"
> t1<-add.species.to.genus(tree,species)
> plotTree(t1,ftype="i")
> species<-"Genus4 sp7"
> t2<-add.species.to.genus(tree,species,where="random")
> plotTree(t2,ftype="i")
> species<-c("Genus1_sp2", "Genus2_sp3", "Genus3_sp2", "Genus4_sp7", "Genus4_sp8", "Genus5_sp3")
> for(i in 1:length(species)) tree<-add.species.to.genus(tree,species[i],where="random")
> plotTree(tree,ftype="i")
If the user supplies a species name for a genus with only one representative in the tree; a species with no congeners in the tree; or a genus that is non-monophyletic, in each case the function will try to do something rational and return a warning.
That's it.



Hi Liam,
ReplyDeleteI am trying to use the add.species.to.genus function, but I am having trouble getting the correct result. The function is only adding the last species of a vector to the phylogeny. Here is my code below and any comment(s) that you can give would be great.
Thanks,
Chris
## (((Frankliniella_tritici:1.0,Frankliniella_occidentalis:1.0):1.0,Echinothrips_americanus:2.0):1.0,Thrips_tabaci:3.0);##
chronotree<-read.tree("test_spp_tre.tree")#load tree
species<-c("Frankliniella_A","Thrips_A","Echinothrips_A")#spp to add to chrono
for(i in 1:length(species)) tree5<-add.species.to.genus(chronotree,species[i],where="root")#add spp to chrono
Hi Chris. This is because your for loop is written incorrectly. It should be:
Deletefor(i in 1:length(species)) chronotree<-add.species.to.genus(chronotree,species[i],
where="root")
plotTree(chronotree)
Otherwise you will not be updating the tree every loop.
- Liam
Thanks Liam, I appreciate your help!
ReplyDeleteIn case other people find it useful, here's a function for adding genus names to the node uniquely containing all members of that genus:
ReplyDeletefor(genus in unique(sub("_.*", "",tree$tip.label))) {
genusRE <- paste("^", genus, "_", sep="")
species<-grep(genusRE,tree$tip.label)
if (length(species)>=2) {
mrca<-findMRCA(tree,tree$tip.label[species])
desc <- getDescendants(tree, mrca)
desc_tips <- desc[desc <= length(tree$tip.label)]
if (length(grep(genusRE, tree$tip.label[desc_tips], invert = TRUE))==0) {
base_node <- mrca - length(tree$tip.label)
if (is.null(tree$node.label[base_node]) || is.na(tree$node.label[base_node])) {
tree$node.label[base_node] = genus
}
}
}
}
Cool!
ReplyDeleteGreat function!
ReplyDeleteWould it be possible to - instead of inserting taxa inside the clade, i.e. above the empirical MCRA - could one define a maximum age above which new taxa could be inserted?
Reason I ask is, if you have a genus that is grossly under-sampled and the few species you have are closely related, the MRCA will be quite young and all inserted taxa will be crammed into this short time-frame, which might be unrealistic.
Thanks!
This comment has been removed by the author.
ReplyDeleteHi Liam,
ReplyDeleteI am working with a Large phylo object that I retrieved from OpenToL. After some edition deleting the "Open Tree of Life Taxonomy identifiers" (ott ids) to leave the species names only, I read the tree in R using "read.newick()" with no problems. Then I tried the add.species.to.genus() with my database, but in doing so I get the following error message:
Error in read.tree(text = write.tree(tree)) :
The tree has apparently singleton node(s): cannot read tree file.
Reading Newick file aborted at tree no. 1
In addition: There were 50 or more warnings (use warnings() to see the first 50)
If I understand well "add.species.to.genus" uses read.tree() to read the tree instead of the read.newick() function, hence singletons in my original tree become a problem at this point. Am I right? If so, what should I do? Should I edit my tree to becorrectly readed by read.tree() first? or there is a way to modify arguments in add.species.to.genus() to use read.newick() instead?
Btw: excelent blog, thanks!
Hmmm. You could try:
Deleteread.tree<-read.newick
## run your code
rm(read.tree)
Not sure, but that might work.
Hi Liam -
ReplyDeleteI've been using this function quite a bit lately, but have run into an issue with my latest dataset. I'm iteratively adding about 20 species from different genera into an ultrametric tree, and for some reason it keeps spitting out a non-ultrametric tree about halfway through the loop, which then leads to a tree with no branch lengths at all in the next iteration. I tried to track it down, and it seems that in some instances, the internal splitTree function returns a subtree with branch lengths that differ just above the default tolerance for 'is.ultrametric', which is also used internally. Thus it thinks the whole tree is non-ultrametric, and adds the new tip without the proper consideration. Do you have any ideas on how to combat this behavior?? It could be simple enough just to specify a slightly larger tolerance on the internal use of is.ultrametric, but I'm not sure how to do that...
Thanks in advance!!
This comment has been removed by the author.
DeleteHi Ryan and Liam,
ReplyDeleteI am having the same problem (If I understood your). I am adding some species in an a tree (that I transformed in ultrametric before), but in some moment the function returns a error message that the tree is not ultrametric.I don't know how I can solve this.
Hi Liam,
ReplyDeleteI was wondering if you had a reply to Ryan and Rodrigo's problem, as I seem to be running into the same issue. Thank you so much for your help.
Dominique
I think that it is probably correct that this error is due to limited precision in the edge lengths of the tree. It can probably be solved by running the function force.ultrametric( ) on your input tree from file after it has been read in. NOTE that this should only be done if the tree is genuinely ultrametric & a deviation from ultrametricity that R detects is just due to branch length rounding.
DeleteIt looks like I did wind up solving this by simply wrapping the function with force.ultrametric() inside my loop.
DeleteGlad to hear it (although I suspect that if you just do this once beforehand it would be sufficient).
DeleteHi everyone,
ReplyDeleteI was wondering if there is a way to add a taxa to a specific node and keeping the tree ultrametric? I also tried the AddTip function{TreeTools} which allows me to correctly add taxa to specific nodes but I don't know how to keep the tree ultrametric when doing it (as default it assigns a zero edge length)?! Any help would be highly appreciated! Thanks a lot!
Best wishes,
Belinda
This comment has been removed by the author.
ReplyDeleteHi Liam,
ReplyDeleteThank you so much for these bunch of codes.
I was wondering how we can do replicate for adding a set of species to the corresponding genus. Since here, one name only is binded to a specific position.
Thank you very much.