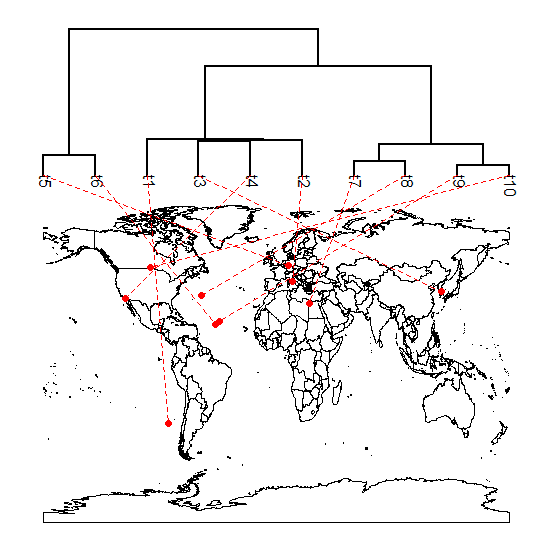
I'm new to geographic maps (see 1, 2), but they are fun to play with so, after a little work, I've added the function phylo.to.map. It now creates an object of class "phylo.to.map" which can be used with the S3 plotting method plot.phylo.to.map. The function also allows optional user control of a bunch of different plotting options; however these are not yet thoroughly documented.
The new version of phytools with these updates (phytools 0.3-19) also depends on the R package maps.
Here's a quick demo:
> require(phytools)
Loading required package: phytools
> packageVersion("phytools")
[1] ‘0.3.19’
> # simulate tree & data
> tree<-pbtree(n=26,scale=100)
> tree$tip.label<-LETTERS[26:1]
> lat<-fastBM(tree,sig2=10,bounds=c(-90,90))
> long<-fastBM(tree,sig2=80,bounds=c(-180,180))
> # now plot projection
> phylo.to.map(tree,cbind(lat,long),asp=1.3)
objective: 168
...
objective: 78
We can also zoom in on particular regions of the map, although that is not well developed. So, for instance:
> tree<-pbtree(n=26,scale=100)
> tree$tip.label<-LETTERS[26:1]
> lat<-fastBM(tree,sig2=10,bounds=c(-36,-16),a=-20)
> long<-fastBM(tree,sig2=80,bounds=c(115,150),a=130)
> # now plot projection
> phylo.to.map(tree,cbind(lat,long),ylim=c(-40,-10), xlim=c(110,155))
objective: 166
...
objective: 114
It's also now possible to project the tree directly onto a geographic map. Users of this method, though, should keep in mind that "ancestral nodes" are just a projection of the tree - and are not intended as a reconstruction of ancestral areas:
> tree<-pbtree(n=26,scale=100)
> tree$tip.label<-LETTERS[26:1]
> tree<-paintSubTree(tree,length(tree$tip)+1,"1")
> lat<-fastBM(tree,sig2=10,bounds=c(-90,90))
> long<-fastBM(tree,sig2=80,bounds=c(-180,180))
> plot.new()
> plot.window(xlim=c(-180,180),ylim=c(-90,90),asp=1.3)
> map("world",add=TRUE)
> phylomorphospace(tree,cbind(long,lat), colors=setNames("red",1),node.by.map=TRUE,add=TRUE, label="horizontal")