Felsenstein (2004; pp. 574-576) gives four different node placements for square phylograms. The most commonly used node placement what Felsenstein calls intermediate, i.e., the node is placed at the midpoint of the uppermost & lowermost edges immediately descended from that node. This is what that looks like for a stochastic 13 taxon tree:
[1] ‘0.4.16’
> plotTree(tree)
Centered node placement puts the node - instead of at the vertical point intermediate between the upper & lowermost immediate daughters - at the midpoint of all tips descended from a node. This is just the average of the upper and lowermost of this set. Here's what that looks like for the same tree:
Like intermediate node placement, weighted node placement uses only the immediate descendants - but it weights their vertical position (inversely) by the edge length leading to each daughter node. E.g.:
(Note that Figure 34.1c in Felsenstein appears to use a slightly different algorithm than that described in the text - which I have followed here - in which internal and tip edges are treated differently.)
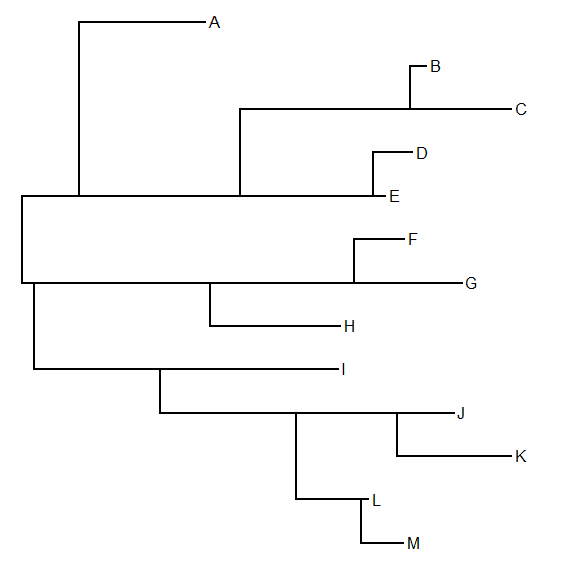
Finally, inner node placement, puts each node in the 'innermost' position of all its descendants. E.g.:
(In this case the algorithm described by Felsenstein (2004; p. 575) seems to be incorrect unless I've misread. I achieved the desired effect not by using the of the y values of the descendant edges; but the vertical position closest to the median vertical position of all the tips.)
All of these methods can also be used with stochastic mapped trees (since plotSimmap is the engine that runs plotTree). For instance:
> plotSimmap(anoletree,nodes="inner",fsize=0.7,ftype="i")
no colors provided. using the following legend:
CG GB TC TG Tr Tw
"black" "red" "green3" "blue" "cyan" "magenta"
> add.simmap.legend(colors=setNames(palette()[1:6],
sort(unique(getStates(anoletree,"tips")))))
Click where you want to draw the legend
That's it. See some of you at 'Evolution' in a few days.




Liam -- hope the Evolution meetings went well. I am embarrassed to hear that in two cases I gave the wrong algorithm for node placement in the book. I will double check and report here what I find.
ReplyDeleteHi Joe.
DeleteYes, it was a fun meeting. Very big. Nearly 2,000 participants, I heard.
I used your algorithm for the 'weighted' node placement, but Figure 34.1c appears to treat nodes leading to terminal edges differently from nodes leading to other internal nodes.
The 'inner' node placement I'm not sure about. I think it may be written down incorrectly in your book, but I may have just misread it, of course.
See you later in the summer. - Liam
I have looked at the book chapter. The algorothms given are, as far as I know correct. But the node placements in Figure 34.1(c) are not correct for the algorithm, at least for the nodes that have two tips as immedite descendants. My Drawgram program must have had a bug -- and may still.
DeleteOops, "alborithms" Anyway thanks for the heads-up about this.
DeleteOops-squared, "algorithms".
ReplyDelete