A phytools user asks:
Does the phylosig function [Ed. for the estimation of phylogenetic signal] work for non-ultrametric trees? Can I include fossils in my phylogeny?
The answer to both these questions is "yes"? With regard to the former inquiry: I can see two main reasons for using a non-ultrametric tree in the computation of phylogenetic signal. The first is that we have some a priori reason to believe that the rate of character evolution differs strongly on different branches of the tree and we want to condition on this rate heterogeneity. The second is that we have extinct taxa (lineages terminating before the present) included in our phylogeny. In both cases we can compute phylogenetic signal for our characters of interest in the usual way.
The second question pertains to including fossil data in the phylogeny. If the user is referring to the practice of including extinct taxa (fossil species) as lineages terminating before the present, then I have already answered the question. However, if what he or she wants to do is attach a datum (or multiple data) to a specific node or to a point along an internal edge, then this is a different matter. Again, it is possible, but a tiny bit more complicated.
The easiest way to do this is by attaching an extra branch of zero length to our tree. If the fossil placement is at an internal node, then we attach the branch to the node as follows (note that I'm going to do this by simulation using phytools::pbtree, but to repeat this, the user has to update phytools to the latest non-CRAN version, v0.1-95, here - also see note at the end of this post):
> library(phytools)
> # first, let's say we have a phylogeny with 20 tips
> tree<-pbtree(n=20)
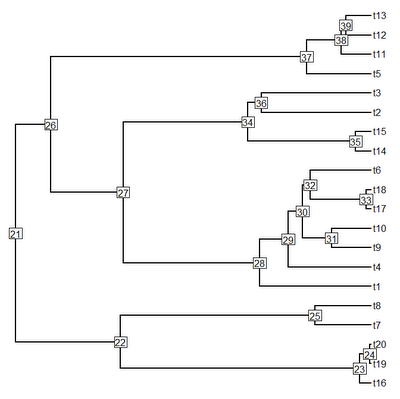
> # plot the tree with node labels
> plotTree(tree,node.numbers=T,pts=F)
> # ok, now let's assume that our fossil data can be
> # confidently placed at, say, node 28
> # first create a new tip
> tip<-list(edge=matrix(c(2L,1L),1,2),tip.label="x1", edge.length=0,Nnode=1L)
> class(tip)<-"phylo"
> # then attach it
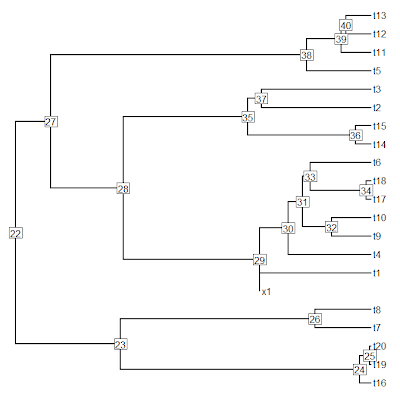
> xtree<-bind.tree(tree,tip,where=28)
> x11(); plotTree(xtree,pts=F,node.numbers=T)
[Note here that all the node numbers have increased by one. This happens even though we have not added any internal nodes to the tree because we have technically added a new tip, and the counting scheme for node numbers begins with number N+1 for N tips in our phylogeny.]
We can repeat this procedure as many times as we want - adding additional fossil phenotypes to the tree.
Normally, then, we would have our data vector containing both the values for all the tip nodes, and the values for ancestral nodes from fossils - the latter named by the convention we have adopted to add new tips to the tree (here, "x1"). Since this is a simulated example, we can just generate some data:
> x<-fastBM(xtree)
> x
t16 t19 ... x1
-3.38357132 -3.04220865 ... 0.17339482
So, you can see that the data for tip species and internal nodes are just treated in exactly the same way in the input data vector. We can then compute phylogenetic signal in the normal way:
> phylosig(xtree,x,method="K")
[1] 1.294228
> phylosig(xtree,x,method="lambda")
$lambda
[1] 0.9967708
$logL
[1] -21.76246
We can do basically the same thing for fossils that we want to place along an edge of the tree, instead of directly at a node. In this case, we would do the following:
> # using our tree from before, say we have a fossil that we
> # want to add halfway between (new) nodes 23 & 26
> tip<-list(edge=matrix(c(2L,1L),1,2),tip.label="x2", edge.length=0,Nnode=1L)
> class(tip)<-"phylo"
> # then attach it
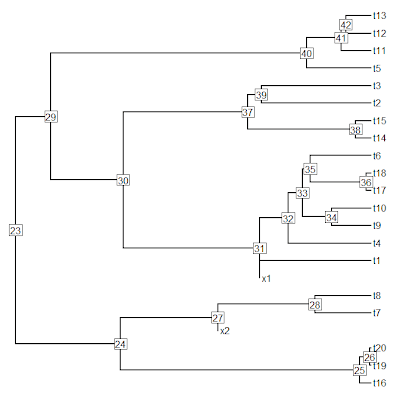
> xtree<-bind.tree(xtree,tip,where=26, position=0.5*xtree$edge.length[which(xtree$edge[,2]==26)])
> plotTree(xtree,pts=F,node.numbers=T)
Then we just proceed as before.
**An important addendum is that the simulation using pbtree for phylogeny simulation does not work with the current CRAN version of phytools, and thus to run the simulation exactly as detailed above, one needs to update to the latest non-CRAN phytools version. This is because of although pbtree simulates trees that are (so far as I can tell) compliant with the ape standard, ape::bind.tree nonetheless finds some problem in how edges are numbered and ordered. This will not result in an error - rather a misplacement of the attached tip in the tree. I have built a temporary fix for this problem into the newest version of pbtree, and I may describe that fix in a separate post.
No comments:
Post a Comment
Note: due to the very large amount of spam, all comments are now automatically submitted for moderation.