Code for the new version of phylomorphospace is here, but because the function calls textxy in the 'calibrate' package internally, and phytools imports from calibrate, users will need to either load calibrate to run phylomorphospace from source, or they can install the newest minor build of phytools (phytools 0.2-24).
Here's a quick demo:
> require(phytools)
Loading required package: phytools
...
> packageVersion("phytools")
[1] ‘0.2.24’
> tree<-pbtree(n=30)
> XX<-fastBM(tree,nsim=2)
> par(mar=c(5.1,4.1,2.1,2.1))
> phylomorphospace(tree,XX) # default
Loading required package: phytools
...
> packageVersion("phytools")
[1] ‘0.2.24’
> tree<-pbtree(n=30)
> XX<-fastBM(tree,nsim=2)
> par(mar=c(5.1,4.1,2.1,2.1))
> phylomorphospace(tree,XX) # default
> phylomorphospace(tree,XX,ylim=c(-3,4),xlim=c(-4.5,3.5), ylab="trait 1",xlab="trait 2")
That's it.

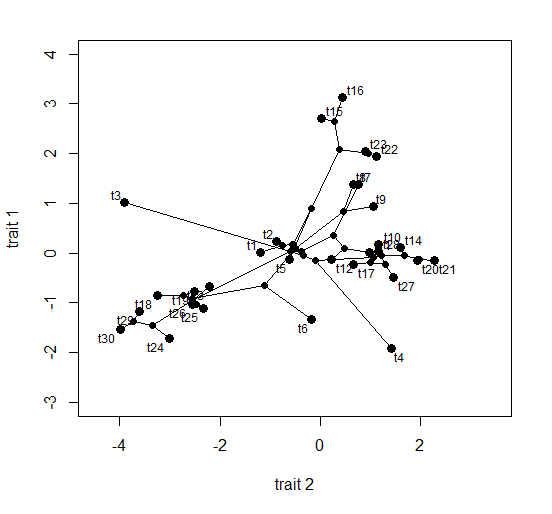
Awesome! Thanks for doing this Liam. It'll be on display Wednesday when I give my continuous evolution tutorial at the Bodega Bay Applied Phylogenetics workshop.
ReplyDeleteIs there a way to change the model of ancestral state reconstruction that phylomorphospace uses?
ReplyDeleteNo - but you can supply.ancestral states that are estimated using another method. This should be done using the argument A which should be a matrix of dimension tree$Nnode x 2, with row names that are the node numbers in the tree. Let me know if you need help with this. - Liam
Delete