A recent user comment asks:
Is there a way to use getDescendents() to color the tips that descend from a certain node (including or excluding the node colors) to use in phylomorphospace()? And if there are several clades one would like to color, how to use the objects of getDescendents for the different clades in the same phylomorphospace?
The answer is "yes" - this is not too hard. (It could be easier, I suspect - but I programmed phylomorphospace a while ago.) Here's how we do it:
> # first let's simulate a tree
> tree<-pbtree(n=30)
> # and data
> XX<-fastBM(tree,nsim=2)
> # plot tree to identify the nodes of the
> # subtrees we want to color
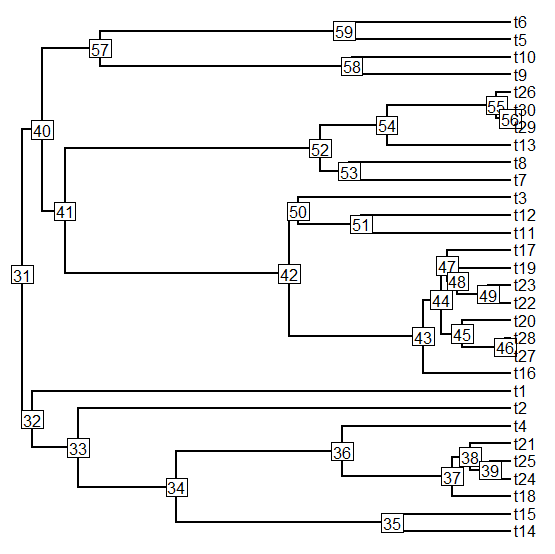
> plotTree(tree,node.numbers=T)
> tree<-pbtree(n=30)
> # and data
> XX<-fastBM(tree,nsim=2)
> # plot tree to identify the nodes of the
> # subtrees we want to color
> plotTree(tree,node.numbers=T)
> # load phangorn for getDescendants
> library(phangorn)
> # now let's say we want to plot nodes
> # descended from "42" red:
> cols<-rep("black",length(tree$tip.label)+tree$Nnode)
> names(cols)<-1:length(cols)
> cols[getDescendants(tree,42)]<-"red"
> # and everything from "36" blue:
> cols[getDescendants(tree,36)]<-"blue"
> # finally, these can even be nested
> cols[getDescendants(tree,45)]<-"yellow"
> # and plot
> phylomorphospace(tree,XX,control=list(col.node=cols),
xlab="X1",ylab="X2")
> library(phangorn)
> # now let's say we want to plot nodes
> # descended from "42" red:
> cols<-rep("black",length(tree$tip.label)+tree$Nnode)
> names(cols)<-1:length(cols)
> cols[getDescendants(tree,42)]<-"red"
> # and everything from "36" blue:
> cols[getDescendants(tree,36)]<-"blue"
> # finally, these can even be nested
> cols[getDescendants(tree,45)]<-"yellow"
> # and plot
> phylomorphospace(tree,XX,control=list(col.node=cols),
xlab="X1",ylab="X2")
If we want to exclude tip labels from this coloring, we have multiple options. We could do the above and then simply set tip labels black:
> cols[1:length(tree$tip)]<-"black"
or we could add an extra line in our assignment, for instance:
> nn<-getDescendants(tree,36)
> nn<-nn[nn>length(tree$tip)]
> cols[nn]<-"blue"
> nn<-nn[nn>length(tree$tip)]
> cols[nn]<-"blue"
That's it!

I'm an idiot. getDescendants is actually a phytools function, so we didn't need to load phangorn.
ReplyDeleteHi Liam!
ReplyDeleteNot sure if I'm doing something wrong but I had several scripts coloring nodes in phylomorphospace and they working great but now the colors are being ignored. I ran this example and I also got no colors on the nodes. Any idea why?
Thanks in advance!
Ricardo
Ricardo. This is a bug. I have fixed it and will post soon. Thanks! Liam
DeleteHere is details of the fix: http://blog.phytools.org/2013/05/bug-fix-update-to-phylomorphospace.html. Let me know if this works. - Liam
DeleteThanks Liam!
Deleteversion 0.6 worked fine
ReplyDelete