In my response to a recent R-sig-phylo message, I posted a trick for assigning the "names" attribute of the vector of edge lengths in a "phylo" object in R such that the name of each edge is given by the formula "parent node #,daughter node #" or, if the daughter node is a tip, then "parent node #,daughter tip label". I'll repeat this hear for the future reference of me & the readers of this blog (if there are any left by the end of this entry):
> library(ape)
> set.seed(100)
> tree<-rtree(10) # simulate random tree
> X<-tree$edge # store tree$edge in a matrix
> # replace all node numbers for tips with their labels
> X[X[,2]%in%1:length(tree$tip),2]<-
tree$tip[X[X[,2]%in%1:length(tree$tip),2]]
> # substitute the names of tree$edge.length for the rows of X
> names(tree$edge.length)<-paste(X[,1],X[,2],sep=",")
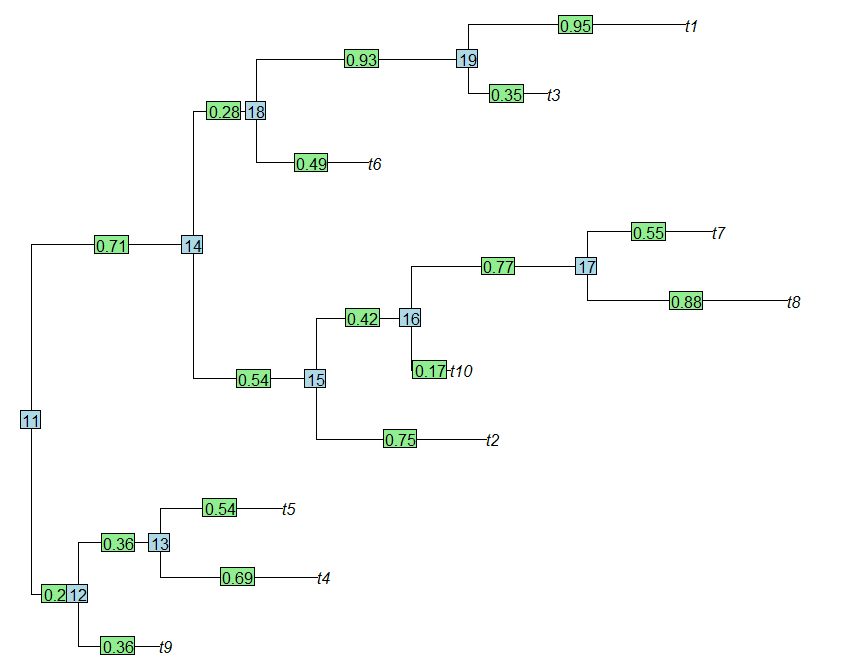
> round(tree$edge.length,2)
11,12 12,t9 12,13 13,t4 13,t5 11,14 14,15 15,t2 15,16
0.20 0.36 0.36 0.69 0.54 0.71 0.54 0.75 0.42
16,t10 16,17 17,t8 17,t7 14,18 18,t6 18,19 19,t3 19,t1
0.17 0.77 0.88 0.55 0.28 0.49 0.93 0.35 0.95
> plot(tree,no.margin=T)
> edgelabels(round(tree$edge.length,2))
> nodelabels()
(We can see that the branch lengths of the tree, plotted as edge labels, line up exactly with the branch names in tree$edge.length. Cool.)
That's it.
I should add to this that we can pull out the lengths of specific branches of the tree as follows:
ReplyDelete> tree$edge.length["18,19"]
18,19
0.9285051
or
> tree$edge.length["15,t2"]
15,t2
0.7489722
The reference and citation generator has vast application according to your needs. This is for those who have interest in the reference generator.
ReplyDelete